 |
 |
- Search
| Clin Exp Reprod Med > Volume 47(4); 2020 > Article |
|
Abstract
Objective
The sperm DNA fragmentation index (DFI) guides the clinician’s choice of an appropriate assisted reproductive technology (ART) procedure. The DFI can be determined using commercially available methodologies, including sperm chromatin dispersion (SCD) kits and sperm chromatin structure assay (SCSA). Currently, when DFI is evaluated using SCD kits, the result is analyzed in reference to the SCSA-derived threshold for the choice of an ART procedure. In this study, we compared DFI values obtained using SCSA with those obtained using SCD and determined whether the difference affects the choice of ART procedure.
Methods
We compared SCSA to two SCD kits, CANfrag (n=36) and Halosperm (n=31), to assess the DFI values obtained, the correlations between tests, the technical repeatability, and the impact of DFI on the choice of ART.
Results
We obtained higher median DFI values using SCD kits than when using SCSA, and this difference was significant for the CANfrag kit (p<0.001). The SCD kits had significantly higher coefficients of variation than SCSA (p<0.001). In vitro fertilization/intracytoplasmic sperm injection (IVF/ICSI) would be chosen for a significantly higher proportion of patients if a decision were made based on DFI derived from SCD rather than DFI determined using SCSA (p=0.003).
Conclusion
Our results indicate that SCD kit-specific thresholds should be established in order to avoid the unnecessary use of IVF/ICSI based on sperm DNA damage for the management of infertility. Appropriate measures should be taken to mitigate the increased variability inherent to the methods used in these tests.
Successful pregnancy depends on several factors, including the integrity of the sperm chromatin, which is represented by the sperm DNA fragmentation index (DFI). An elevated DFI is inconducive to both fertilization and pregnancy [1-8]. Clinicians therefore recognize the value of DFI in the evaluation of male infertility in couples with recurrent pregnancy failure and in the choice of an appropriate assisted reproductive technology (ART) procedure. A number of methodologies are currently available to assess DFI. Two of these detection methods are based on the denaturing capacity of sperm chromatin: sperm chromatin structure assay (SCSA) and sperm chromatin dispersion (SCD) kits.
SCSA, as the gold standard for the assessment of sperm DNA fragmentation, consists of a fixed flow cytometry protocol, requires a proprietary software program (SCSAsoft; SCSA Diagnostics, Brookings, SD, USA) and produces a highly repeatable measure of DFI [9]. SCD kits are technician-dependent light microscope tests that measure 50–500 sperm per sample to provide a DFI based on the presence or absence of a dispersion halo around the fragmented or non-fragmented sperm, respectively [10]. Several SCD kits are available and serve as inexpensive alternatives to SCSA.
The proprietors of SCSA have clasd statistical categories of sperm fertility potential based on DFI, with ≤15% considered to indicate excellent to good fertility potential, 15%–25% good to fair fertility potential, >25% fair to poor fertility potential, and >50% very poor integrity. The probability of a successful pregnancy outcome sharply declines with a DFI >25% when female factor infertility is excluded [7,11,12], and the suggested clinical intervention when DFI is >25% is in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI) rather than in vivo or intrauterine insemination [13-15]. Similar to SCSA, SCD has been indicated in published studies to have a predictive threshold between 20% and 27% for infertile men [16-19]. A threshold above 17%–18% has been found to affect the fertilization outcome [18,20]. This cutoff is specific to one SCD kit, Halosperm, and is not a global threshold that is applicable across all SCD kits. Additionally, no general consensus exists regarding the threshold above which a certain ART procedure should be selected based on DFI from determined with a particular SCD kit, and the current standard is to use SCSA thresholds. To add another layer of complexity, studies comparing DFI generated by SCSA and SCD have indicated both concordance between the 2 tests and discordance, such as a higher DFI obtained when an SCD kit is utilized [16,17,21,22].
In this study, we compared SCSA to the two most commonly used SCD kits (CANfrag and Halosperm) with regard to the DFI values obtained and the technical repeatability. Our goal was to determine whether the values generated from each of these kits would be similar in the same patient. We also investigated whether the type of kit used and the DFI generated affects the type of ART procedure chosen for the patient.
A total of 41 male patients (age range, 27–45 years) were enrolled in this study, which was performed at the Andrology Centre in Coimbatore City, Tamil Nadu, India. Semen samples were collected from each patient after informed consent was obtained. All procedures were performed according to Institutional Review Board policy. All patients underwent serology assessment for viral or bacterial infections, including tests for human immunodeficiency virus (HIV)-1 and 2, hepatitis B surface antigen, hepatitis C virus, and other standard laboratory tests for sexually transmitted infections, before undergoing the SCSA or SCD tests. Prior to sample collection, patients were asked to adhere to an ejaculatory abstinence regime spanning 24–48 hours. The semen samples were collected via masturbation into a sterile wide-mouthed calibrated container. After liquefaction for 1 hour at room temperature, 200–500 µL of the raw semen was aliquoted into cryovials without cryoprotectant and flash-frozen in liquid nitrogen. Samples were analyzed fresh or frozen/thawed. Two SCD kits (CANfrag and Halosperm) and SCSA were chosen for the DFI analyses. A CANfrag kit was utilized on 36 patient samples and a Halosperm kit on 31 samples, and all 41 patients were assessed via SCSA (Figure 1). The DFI value, the correlation of DFI between the different assays, the coefficient of variation (CV) between technical replicates, and the impact on the choice of ART procedure were compared among SCSA and the specific kits (Figure 1). All reagents were purchased from Sigma-Aldrich (Millipore-Sigma, St. Louis, MO, USA) unless otherwise noted.
Individual semen samples, stored in liquid nitrogen tanks (−196°C), were thawed in a 37°C water bath and then immediately placed on crushed ice. An aliquot of raw semen was transferred to a solution of TNE buffer (0.01 M Tris-HCl, 0.15 M NaCl, and 1 mM EDTA; pH 7.4) at 4°C to yield a final concentration of approximately 1–2×106 sperm/mL. A total of 200 µL of this sperm suspension was admixed with 400 µL of a solution containing 0.08 N HCl, 0.15 M NaCl, and 0.1% (v:v) Triton X-100 at 4°C. Importantly, the HCl was diluted from a commercial solution of 2.0 N HCl. After 30 seconds, sperm were stained by adding 1.2 mL of staining solution containing 6 µg/mL acridine orange (AO, chromatographically purified; Polysciences, Warrington, PA, USA), 0.2 M Na2PO4, 0.1 M citric acid (pH 6.0), 1 mM EDTA, and 0.15 M NaCl to yield an AO:DNA-P molar ratio of ≥2 [23]. The acid-/AO-stained sample was placed in a flow cytometer (FACSCalibur; BD Biosciences, San Jose, CA, USA) sample chamber, and sample flow was initiated to bring the sheath flow and sample flow to equilibrium within 2 minutes. Subsequently, 5,000 sperm were analyzed at an event rate of 100–250 events/sec. If the event rate exceeded 250 events/sec, a new sample was prepared to ensure full equilibrium between the AO dye and the sperm. The flow cytometer was calibrated with a reference sample at the start of sample analysis, and the same reference sample was analyzed after every five test samples to calibrate the instrument. Each test sample was analyzed in duplicate, and replicates of the data were utilized to determine the percentage of sperm with measurably increased red fluorescence (sperm with fragmented DNA as determined using SCSAsoft). If a >10% difference in DFI was observed between the raw X and Y means in the replicates, the sample was repeated. The standard deviations between the replicates were calculated.
Low-melting-point agarose, which was pre-provided in a microcentrifuge tube, was placed in a float in boiling water (90°C–100°C) for 5 minutes and then transferred to a 37°C water bath for equilibration. An aliquot of fresh or flash-frozen semen sample was added to this tube of melted agarose in order to achieve a final sperm concentration of 15–20 million/mL. A volume of 150 µL of this agarose-semen sample mixture was pipetted onto the provided slide, covered with a coverslip (25 mm×50 mm), and allowed to solidify at 4°C for 5 minutes. The coverslip was carefully removed, and the slide was kept on a horizontal staining tray and immersed in 1 mL of acid denaturant solution for 7 minutes. After this period, the excess acid solution was drained, 1 mL of lysis buffer was added, and the slide was incubated at room temperature for 10 minutes. The slide was washed gently with 20 mL of distilled water to remove the lysis buffer, and sequential dehydration steps were performed using dehydrating solutions 1, 2, and 3 (provided in the kit). The slides were air-dried and stained using the staining solution provided with the kit (working stain prepared: 300 µL of stain+100 µL of dilution buffer), 250 µL of which was used to stain each slide. The slides were incubated for 5 minutes, air-dried, and examined under ×400 magnification using a light microscope (CH20i; Olympus, Tokyo, Japan). Each slide was scored for 200 spermatozoa by three lab technicians. Sperm cells with absent or small halos (≤1/3 of the head width) were counted as sperm with fragmented DNA; otherwise, they were considered normal sperm. The means and standard deviations were calculated.
Low-melting-point agarose (1%), which was pre-provided in a microcentrifuge tube, was placed in a float in boiling water (90°C–100°C) for 5 minutes and then transferred to a 37°C water bath for equilibration. An aliquot of fresh or flash-frozen semen sample was added to this tube of melted agarose in order to achieve a final sperm concentration of 5–10 million/mL. A drop of 10–15 µL of this agarose-semen sample mixture was pipetted onto the pre-treated slide and covered with a coverslip (18×18 mm or 22×22 mm), the edges of which were pressed gently to obtain an uniform distribution of the gel on the slide. The slide was then incubated at 4°C for 5 minutes. The coverslips were carefully removed, and the slides were immediately placed on a horizontal staining tray. An acid denaturant solution was freshly prepared (80 µL of the acid denaturant+10 mL of distilled water, provided in the kit), added to the gel, and allowed to react for 7 minutes. On completion, the slides were placed on another tray of lysis buffer and incubated at room temperature for 25 minutes. The slides were washed with abundant distilled water to completely remove the lysis solution and were then incubated for 5 minutes. Following this, the slides were sequentially dehydrated using 70%, 90%, and 100% ethanol (Changshu Hongsheng Fine Chemical, Jiangsu, China) for 2 minutes each, respectively. The slides were air-dried and stained using Diff-Quik Stain (Cell Life Ref: CL06; Cell Life, Visakhapatnam, India). First, azure A (eosin-red) stain was added and incubated for 7 minutes, and azure B (nigrosin-blue) stain was added and incubated for 7 minutes. The slides were incubated for an additional 5 minutes, air-dried, and examined under ×400 magnification using a light microscope (CH20i, Olympus). Each slide was scored for 500 spermatozoa by three lab technicians. Sperm cells with absent or small halos (≤1/3 of the head width) were counted as sperm with fragmented DNA; otherwise, they were considered normal sperm. The means and standard deviations were calculated.
The CV was calculated for the triplicate readings from SCD and the duplicate readings from SCSA using the formula ([standard deviation/mean]×100). The DFI value was used as an indicator of sperm quality, namely excellent (<15%), good to fair (15%–25%), fair to poor (>25%–50%) and very poor (>50%). The specific statistical tests used to determine significant differences (p<0.05) are mentioned in the respective parts of the Results section.
The DFI derived from SCSA was significantly lower than the DFI derived using the CANfrag SCD kit (Wilcoxon test, p<0.001) (Table 1). Additionally, in the correlation analysis between SCSA and the SCD kits, both SCD kits had a strong significant correlation with the SCSA kit (Spearman rank tests, p-values in Table 1). Of the patients tested, 29 had their sperm analyzed using all three tests. Two-way analysis of variance (ANOVA) revealed that DFI was the lowest when determined using SCSA and was slightly greater when determined using either Halosperm or CANfrag (Figure 2). The median DFI levels were 19.3%, 24.7%, and 29% for SCSA, Halosperm and CANfrag, respectively, and the level associated with CANfrag was significantly higher than those for SCSA or Halosperm (two-way ANOVA, p<0.0001) (Figure 2).
We calculated the CVs of the replicates for each patient to determine the technical repeatability of the tests. The CVs ranged from 0%–18% (mean, 4.7%) for SCSA, 2%–54% (mean, 24.2%) for CANfrag and 7%–62% (mean, 27.2%) for Halosperm. The CV associated with SCSA was significantly lower than the CVs associated with CANfrag or Halosperm (repeated-measures ANOVA and Bonferroni post-hoc test, p<0.001). When an arbitrary cut-off value of 10% was set for CV, we observed that 30 of 36 patients (83.3%) for CANfrag, 29 of 31 (93.5%) for Halosperm, and 4 of 41 (9.7%) for SCSA would require repetition of the test. The mean DFI, along with its 95% confidence interval, was calculated for each test (Figure 3). Outside lines closer to the mean indicate higher repeatability of the test.
Since DFI informs the choice of ART procedure, we compared DFI and the subsequent ART procedure chosen in 29 patients when SCSA, Halosperm or CANfrag was used. A heatmap representing the classification of the sperm into four categories of sperm potential showed a striking difference in the number of patients presenting with a higher DFI and lower fertility potential when an SCD kit was used (Figure 4A). The clinical management plan for each of these patients was further analyzed. This comparison revealed that if the SCSA testing method had been used for assessing sperm damage, and 25% DFI was set as the cutoff above which IVF/ICSI was selected instead of in vivo or intrauterine insemination, clinicians would have selected only nine of 29 patients for IVF/ICSI when using SCSA as opposed to 14 of 29 and 22 of 29 when using Halosperm and CANfrag, respectively (chi-square test, p=0.003) (Figure 4B).
In this proof-of-concept report, we established that commercially available SCD kits showed higher DFI values than SCSA for the same patient, were technically less reliable, and resulted in the ART approach of IVF/ICSI being chosen for more patients. This is the first study of its kind that not only included the analysis of DFI across two different methodologies (SCSA and SCD) but also employed the use of various SCD kits on the same patient sample, highlighting the high subjectivity and variability of clinical DFI values for the same patient across various testing methods.
In SCSA, the use of flow cytometry is employed to determine the levels of intact native double-stranded DNA and fragmented single-stranded DNA. One of the major advantages of flow cytometric assessments is the number of cells analyzed; in this case, a minimum of 5,000 individual sperm cells are analyzed in duplicate. The use of a proprietary software program to convert the data into a DFI value removes the ambiguity of an operator-dependent method and provides a robustness that is lacking in other methodologies. In comparison, the SCD kit methods involve the analysis of 100–500 cells in select fields, depends on the experience of technicians, and provides an incomprehensive representation of the DNA integrity of the complete sample. It is imperative to choose an appropriate method with strict guidelines when using sperm DNA integrity reports to inform the clinical management of infertile couples.
It is currently challenging for researchers and clinicians to compare DFI derived from different assays across diverse studies to assess the risk of male factor infertility. In our results, a higher DFI was recorded when using a SCD kit than when using SCSA, and a similar result has been found in other studies [20,24,25]. The results regarding CV and technical repeatability appear to be contentious; the Halosperm pioneer laboratory reported a CV of 6%–12% [10], which is much lower than our observed values (2%–54% for CANfrag and 7%–62% for Halosperm). The results of our study also suggest that SCSA produced reliable results with duplicates in >90% of cases, while using an SCD test produced reliable results in only <17% of cases. A limitation of this study is the lack of clinical data and the need for validation in a much larger patient population with known clinical outcomes. Regardless, our manuscript brings into focus the subjective nature of SCD and emphasizes that it is unrealistic to expect technicians of varying levels of experience to be sufficiently skilled to assess DFI accurately. The availability of several SCD test kits also confounds the accurate interpretation of results [26]. Ideally, the proprietors of SCD kits should put forth a cut-off value specific to each kit, as in the case of SCSA (i.e., >10% DFI between technical replicates), above which the test needs to be repeated. This is currently unaddressed, and no guidelines are available for use by inexperienced technicians or labs.
The majority of andrology laboratories that use SCD test kits currently use the available SCSA thresholds to determine sperm quality and inform subsequent clinical management. Our analyses of the impact of DFI derived from SCD versus that derived from SCSA indicated that when SCD kits are used, a significantly higher number of patients would be categorized as having poor-quality sperm, and subsequently, a higher number of patients would be selected to undergo IVF/ICSI, creating ambiguity in the clinical setting. Given that a higher DFI is known to be derived from SCD kits, it is critical to utilize threshold values specific to the patient population or infertility center for the preferred SCD kit in order to maximize the potential derived from each of these tests, as suggested by Ribas-Maynou et al. [20]. This approach allows refinement of the choice of ART procedure based on SCD results and avoids the unwarranted use of ICSI for the clinical management of male factor infertility. As in the case of technical repeatability, thresholds to simplify patient management are also needed. This is achievable only when a larger number of patients with known clinical outcomes are compared with regard to DFI values obtained using a specific SCD test and the existing gold-standard assay (SCSA). Additionally, a threshold value must be defined not only for male factor infertility but also to identify a cutoff DFI above which an increased risk of miscarriage or pregnancy failure is observed.
In conclusion, our report establishes that SCSA is reliable with respect to technical repeatability and provides a more streamlined approach for the management of infertile couples. Parameters must be established for a chosen SCD test instead of utilizing the thresholds set for SCSA for the clinical management of infertile couples.
Acknowledgments
We thank the patients who participated in this study. We would also like to acknowledge Professor Don Evenson for his valuable input.
Notes
Figure 1.
Flowchart of study design. A total of 41 patients were enrolled in the study, all of whom were assessed via sperm chromatin structure assay (SCSA). Subsets of patients were also compared using CANfrag (n=36) or Halosperm (n=31) sperm chromatin dispersion kits as indicated. The DNA fragmentation index (DFI), correlation, % coefficient of variation (CV), and impact of the derived DFI on the clinical decision were assessed.

Figure 2.
Lower DNA fragmentation index (DFI) values obtained with sperm chromatin structure assay (SCSA) than with sperm chromatin dispersion (SCD) kits. The graph indicates the DFI with the line within the box set at the median in 29 semen samples analyzed using SCSA (median DFI, 19.3%), Halosperm (median DFI, 24.7%) or CANfrag (median DFI, 29%). The DFI of SCSA and Halosperm was significantly lower than CANfrag. a)p<0.001, Statistically.
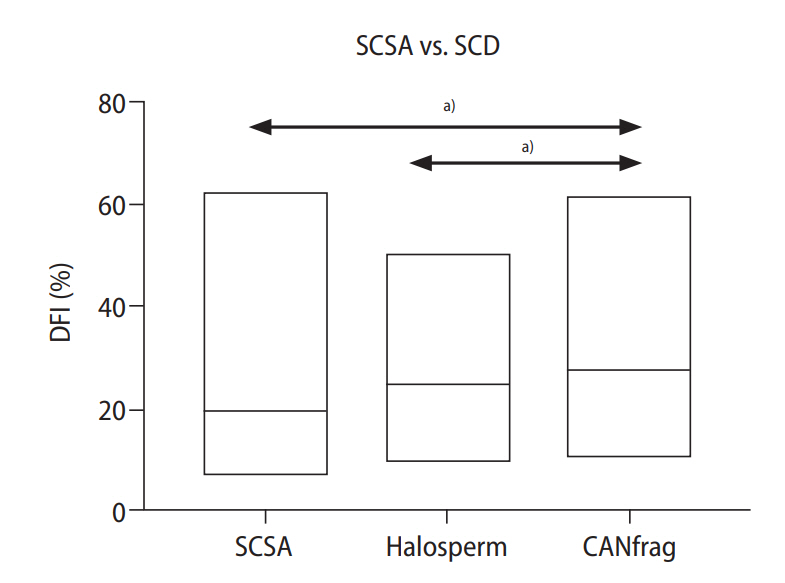
Figure 3.
Technical repeatability of the test. The graph indicates the mean DNA fragmentation index (DFI; black line) and the 95% confidence interval (green, upper limit; red, lower limit) of the different tests. (A) Sperm chromatin structure assay (SCSA). (B) Halosperm. (C) CANfrag.

Figure 4.
(A) Differences in categorizing sperm potential. A heat map of 29 patients with the representative DNA fragmentation index (DFI) for a patient when sperm chromatin structure assay (SCSA), Halosperm, or CANfrag were used. The color-coding represents the categories of sperm potential established for SCSA (green, <15% DFI, indicating excellent fertility potential; yellow, 15%–25% DFI, indicating good to fair fertility potential; orange, >25%–50% DFI, indicating fair to poor fertility potential; red, >50% DFI, indicating very poor integrity). (B) Clinical management is affected by the type of assay utilized to measure the DFI. The bars provide a visual representation of the number of patients selected for in vitro fertilization/intracytoplasmic sperm injection (IVF/ICSI; in red) or in vivo/intrauterine fertilization (in vivo/IUI) when SCSA, Halosperm, or CANfrag kits were used. The chi-square test revealed a statistically significant difference in the type of assisted reproductive technology (ART) procedure chosen based on the assay (p=0.003). SCD, sperm chromatin dispersion. a)Statistically significant.

References
1. Bareh GM, Jacoby E, Binkley P, Chang TC, Schenken RS, Robinson RD. Sperm deoxyribonucleic acid fragmentation assessment in normozoospermic male partners of couples with unexplained recurrent pregnancy loss: a prospective study. Fertil Steril 2016;105:329-36.


2. Bungum M, Humaidan P, Axmon A, Spano M, Bungum L, Erenpreiss J, et al. Sperm DNA integrity assessment in prediction of assisted reproduction technology outcome. Hum Reprod 2007;22:174-9.



3. Dar S, Grover SA, Moskovtsev SI, Swanson S, Baratz A, Librach CL. In vitro fertilization-intracytoplasmic sperm injection outcome in patients with a markedly high DNA fragmentation index (>50%). Fertil Steril 2013;100:75-80.


4. Leach M, Aitken RJ, Sacks G. Sperm DNA fragmentation abnormalities in men from couples with a history of recurrent miscarriage. Aust N Z J Obstet Gynaecol 2015;55:379-83.


5. Micinski P, Pawlicki K, Wielgus E, Bochenek M, Tworkowska I. The sperm chromatin structure assay (SCSA) as prognostic factor in IVF/ICSI program. Reprod Biol 2009;9:65-70.


6. Oleszczuk K, Giwercman A, Bungum M. Sperm chromatin structure assay in prediction of in vitro fertilization outcome. Andrology 2016;4:290-6.


7. Spano M, Bonde JP, Hjollund HI, Kolstad HA, Cordelli E, Leter G. Sperm chromatin damage impairs human fertility. The Danish First Pregnancy Planner Study Team. Fertil Steril 2000;73:43-50.


8. Virro MR, Larson-Cook KL, Evenson DP. Sperm chromatin structure assay (SCSA) parameters are related to fertilization, blastocyst development, and ongoing pregnancy in in vitro fertilization and intracytoplasmic sperm injection cycles. Fertil Steril 2004;81:1289-95.


9. Evenson DP, Darzynkiewicz Z, Melamed MR. Relation of mammalian sperm chromatin heterogeneity to fertility. Science 1980;210:1131-3.


10. Fernandez JL, Muriel L, Rivero MT, Goyanes V, Vazquez R, Alvarez JG. The sperm chromatin dispersion test: a simple method for the determination of sperm DNA fragmentation. J Androl 2003;24:59-66.


11. Evenson DP, Jost LK, Marshall D, Zinaman MJ, Clegg E, Purvis K, et al. Utility of the sperm chromatin structure assay as a diagnostic and prognostic tool in the human fertility clinic. Hum Reprod 1999;14:1039-49.



12. Simon L, Zini A, Dyachenko A, Ciampi A, Carrell DT. A systematic review and meta-analysis to determine the effect of sperm DNA damage on in vitro fertilization and intracytoplasmic sperm injection outcome. Asian J Androl 2017;19:80-90.


13. Evenson DP. Evaluation of sperm chromatin structure and DNA strand breaks is an important part of clinical male fertility assessment. Transl Androl Urol 2017;6(Suppl 4): S495-500.



14. Evenson DP, Wixon R. Data analysis of two in vivo fertility studies using Sperm Chromatin Structure Assay-derived DNA fragmentation index vs. pregnancy outcome. Fertil Steril 2008;90:1229-31.


15. Gosalvez J, Lopez-Fernandez C, Fernandez JL. Sperm chromatin dispersion test: technical aspects and clinical applications. In: Zini A, Agarwal A, editors. Sperm chromatin dispersion test: technical aspects and clinical applications. New York: Springer; 2011. p. 151-70.
16. Chohan KR, Griffin JT, Lafromboise M, de Jonge CJ, Carrell DT. Comparison of chromatin assays for DNA fragmentation evaluation in human sperm. J Androl 2006;27:53-9.


17. Fernandez JL, Muriel L, Goyanes V, Segrelles E, Gosalvez J, Enciso M, et al. Simple determination of human sperm DNA fragmentation with an improved sperm chromatin dispersion test. Fertil Steril 2005;84:833-42.


18. Nunez-Calonge R, Caballero P, Lopez-Fernandez C, Guijarro JA, Fernandez JL, Johnston S, et al. An improved experimental model for understanding the impact of sperm DNA fragmentation on human pregnancy following ICSI. Reprod Sci 2012;19:1163-8.


19. Velez de la Calle JF, Muller A, Walschaerts M, Clavere JL, Jimenez C, Wittemer C, et al. Sperm deoxyribonucleic acid fragmentation as assessed by the sperm chromatin dispersion test in assisted reproductive technology programs: results of a large prospective multicenter study. Fertil Steril 2008;90:1792-9.


20. Ribas-Maynou J, Garcia-Peiro A, Fernandez-Encinas A, Abad C, Amengual MJ, Prada E, et al. Comprehensive analysis of sperm DNA fragmentation by five different assays: TUNEL assay, SCSA, SCD test and alkaline and neutral Comet assay. Andrology 2013;1:715-22.


21. Kunkitti P, Sjodahl A, Bergqvist AS, Johannisson A, Axner E. Comparison of DNA fragmentation assay in frozen-thawed cat epididymal sperm. Reprod Domest Anim 2016;51:618-22.


22. Ortiz I, Urbano M, Dorado J, Morrell JM, Al-Essawe E, Johannisson A, et al. Comparison of DNA fragmentation of frozen-thawed epididymal sperm of dogs using Sperm Chromatin Structure Analysis and Sperm Chromatin Dispersion test. Anim Reprod Sci 2017;187:74-8.


23. Darzynkiewicz Z, Traganos F, Sharpless T, Melamed MR. Thermal denaturation of DNA in situ as studied by acridine orange staining and automated cytofluorometry. Exp Cell Res 1975;90:411-28.


24. Javed A, Talkad MS, Ramaiah MK. Evaluation of sperm DNA fragmentation using multiple methods: a comparison of their predictive power for male infertility. Clin Exp Reprod Med 2019;46:14-21.




- TOOLS







