 |
 |
- Search
| Clin Exp Reprod Med > Volume 47(4); 2020 > Article |
|
Abstract
Objective
The goal of the present study was to investigate the rate of chromosomal aneuploidies in surplus embryos after sex determination at the cleavage stage. Then, the same chromosomal aneuploidies were evaluated in blastocysts after extended culture.
Methods
Sixty-eight surplus embryos were biopsied at the cleavage stage and incubated for an additional 3 days to allow them to reach the blastocyst stage. The embryos were reanalyzed via fluorescence in situ hybridization (FISH) to examine eight chromosomes (13, 15, 16, 18, 21, 22, X, and Y) in both cleavage-stage embryos and blastocysts.
Results
Although the total abnormality rate was lower in blastocysts (32.35%) than in cleavage-stage embryos (45.58%), the difference was not significant (p=0.113). However, when we restricted the analysis to autosomal abnormalities, we observed a significant difference in the abnormality rate between the cleavage-stage embryos (44.11%) and the blastocysts (17.64%, p=0.008). A higher rate of sex chromosomal abnormalities was also observed in cleavage-stage embryos (29.4%) than in blastocysts (14.70%, p=0.038).
The assessment of morphology alone is not an adequate method of differentiating euploid embryos from aneuploid embryos to facilitate a viable pregnancy and a live birth [1,2]. Therefore, an alternative approach is required for precise embryo selection; this approach should include genetic and cytogenetic assessments following morphological investigation for the detection of aneuploidy [1]. Chromosomal abnormalities can be independent of the morphological features of the embryo. These abnormalities are widespread during meiosis and mitosis early in embryonic development, and they result in aneuploidies, such as mosaicism, in embryos [3]. Chromosomal instability causes a marked decline in the success rate of conception [4]. Genotype determination using cytogenetic techniques accompanied by assessment of the morphology of the embryo has been demonstrated to significantly improve both implantation and live birth rates [5]. The prevalence of aneuploidy has been reported to be greater than 50% of preimplantation embryos in in vitro fertilization programs [3,6-9]. Aneuploidy is a pivotal genetic condition that can lead to implantation failure, miscarriage, and congenital abnormalities and impacts the success rates of assisted reproductive technology programs [10]. It has been reported that, in cases of advanced maternal age, almost 40% of aneuploid embryos with chromosomal abnormalities can reach the blastocyst stage with similar morphological appearance to euploid embryos [1,7].
Several studies have shown that multiple chromosomes (13, 15, 16, 18, 21, 22, X, and Y) are involved in the high rates of aneuploidies in cleavage-stage embryos. These aneuploidies include trisomies that cause spontaneous abortion, along with those in pregnancies carried to term in the form of trisomic syndrome [11,12]. In addition, mosaicism increases the risk for intrauterine fetal demise or uniparental disomy [13,14]. Some studies have declared that mosaic embryos self-correct via a different aneuploidy rescue mechanism from non-mosaic aneuploid embryos. Following such a mechanism, the aneuploid cells are excluded and do not participate in the embryo’s cytogenetic status [1,2,4]. Recently, Fesahat et al. [15] reported a significant difference in the aneuploidy rates between sex chromosomes and autosomal chromosomes in human cleavage-stage embryos. However, according to a previous study, biopsies are only performed at the blastocyst stage in about 7% of cycles, up from less than 1% previously [16]. Almost 67% of biopsies were performed at the cleavage stage using fluorescence in situ hybridization (FISH).
The objective of the present study was to determine the common abnormalities (aneuploidies) in embryos resulting from intracytoplasmic sperm injection (ICSI) in two steps. The first step was the evaluation of autosomal abnormalities in biopsies taken for sex selection in the second round of FISH. The second step was the evaluation of autosomal abnormalities in blastocysts generated from surplus embryos from a sex selection program.
The study was approved by the independent Ethics Committee at the Yazd Reproductive Sciences Institute (IR.SSU.REC.1396.20). Written informed consent was obtained from all participants in the study. All experiments were performed in accordance with relevant guidelines and regulations.
Sixty-eight surplus embryos from 37 young couples (aged 25–35 years) with proven fertility were subjected to sex chromosome analysis. The surplus embryos were analyzed in two sequential rounds. The first round included rewashed sex selection slides from surplus embryos, which developed to reach the blastocyst stage. Next, the surplus embryos were assessed in the blastocyst stage (Figure 1).
This work was done at Avicenna Infertility Center in Tehran between August and November 2018. The inclusion criteria were: surplus embryos from sex selection that had reached the blastocyst stage, normal ejaculation according to 2010 World Health Organization criteria, and women <40 years old without a history of recurrent pregnancy loss or genetic disorders. All participants had at least one child.
After ICSI, the injected oocytes were cultured in 20-μL drops of cleavage medium (ORIGIO Sequential Cleav medium; Origio, Måløv, Denmark) covered with paraffin oil. Fertilization was verified 16–18 hours after ICSI. Then, the fertilized oocytes were cultured in cleavage medium for 48 hours, and embryonic development was assessed according to the categorization system proposed by Gardner [19,20]. On day 3, the cleavage-stage embryo biopsy was performed for the genetic analysis via FISH. Genetic testing was carried out via laser biopsy on 2 blastomeres from each top-quality embryo that had at least 7 blastomeres available for sex selection analysis [19]. After the blastomeres were lysed and fixed on 2 different glass slides. The nuclei were analyzed using the FISH method (Figure 1) [7].
Reanalysis slides from surplus embryos were subjected to sex selection for autosomal chromosomes (13, 15, 16, 18, 21, and 22) via the FISH method. The procedure included washing in phosphate-buffered saline, fading of the previous probe, and a subsequent dehydration series in ethanol. Finally, the slides were hybridized with a second set of probes. Two slides were applied for the chromosomal X series (13, 18, 21, and X) (Cytocell; Oxford Gene Technology, Oxfordshire, UK) and Y series (15, 16, 22, and Y) centromeric probes (MetaSystems GmbH, Altlussheim, Germany). Then, the slides were assessed using a fluorescent microscope (Olympus BX51 and Genetics GSL-10 with BX61; Olympus, Tokyo, Japan). The installation filter sets on the microscope were triple-band filters (aqua, orange, and green) and single-band pass filters (red, green, and aqua). Images were captured at ×60 or ×100 magnification using imaging software (Spectral Instruments Imaging, Tucson, AZ, USA).
The blastocysts were graded according to the categorization system proposed by Gardner [19,20]. On day 3, the blastocyst biopsy of the surplus embryos was conducted for sex selection with assessment of the autosomal and sex chromosomes (13, 15, 16, 18, 21, 22, X, and Y) via the FISH method. Blastocyst biopsy of trophectoderm (TE) cells was performed on day 5 after fertilization from hatching blastocysts [21]. For the biopsy, each embryo was placed in 5 µL of blast medium (ORIGIO Sequential Blast) under mineral oil in a separate dish. The blast medium was warmed in an incubator with carbon dioxide. The hatching blastocyst was left in the blast medium instead of Ca2+-free medium for the biopsy. This prevented the loosening of the adhesion junction between cells and avoided mixing of TE and inner cell mass (ICM) cells. TE cells were gently captured by a hooked (30°) micropipette. Then, 5–20 TE cells were cutoff with a noncontact 2.3-μm laser (ZILOS-tk; Hamilton Thorne, Beverly, MA, USA) in the process of laser-assisted hatching. The blastocyst was then released from the holding micropipette. Finally, the hook micropipette was very gently rubbed against the holding micropipette to facilitate the release of biopsied cells from the hook micropipette.
The separated TE cells were placed in droplets of hypotonic medium that had already been warmed in the incubator with carbon dioxide for 10–30 minutes. The duration of incubation depended on the number of TE cells being biopsied. Then, swelling cell samples were transferred into a scored circle on a coated slide and fixed with cold Carnoy solution (methanol:acetic acid, 3:1). Finally, the slides were left to air-dry at room temperature.
The slides prepared from each embryo for the FISH testing for the chromosome X series (13, 18, 21, and X; Oxford Gene Technology) and Y series (15, 16, 22, and Y) centromeric probes (MetaSystems, Altlussheim, Germany) were checked with fluorescent microscopy. The FISH procedure was accomplished according to modified instructions (Oxford Gene Technology).
SPSS ver. 16.0 (SPSS Inc., Chicago, IL, USA) was used for data analysis.T he data were categorical and were expressed as percentages. The chi-square and Fisher exact tests were used for data analysis as appropriate. The p-values ≤0.05 were considered to indicate statistical significance.
A significant rescue in autosomal aneuploidy was observed in blastocysts relative to cleavage-stage embryos (17.64% vs. 44.11%, respectively; p=0.008). Significant self-correction of sex abnormalities was also observed in blastocysts relative to cleavage-stage embryos (14.70% vs. 29.4%, respectively; p=0.038). In addition, significant differences were seen in the aneuploidy rates in cleavage-stage embryos compared to blastocysts in chromosomes 13 (20.4% vs. 0%, respectively; p<0. 001), 21 (20.5% vs. 0%, respectively; p<0.001), 16 (26.4% vs. 11.76%, respectively; p=0.029), and Y (30.8% vs. 8.82%, respectively; p=0.001). The rate of chaotic aneuploidy in blastocysts was 13.23%, the frequency of polyploidy was 7.35%, and the rate of mosaicism in TE cells was 14.70% (Figure 2). Overall, no significant differences were found between the total abnormality of embryos in the cleavage and blastocyst stages (45.58% vs. 32.35%, respectively; p=0.113). The numbers of abnormalities in each group are also presented separately in Table 1.
The aim of the current study was to estimate the rates of autosomal aneuploidies in surplus cleavage-stage embryos generated from young, fertile women admitted to a sex selection program. Moreover, the rates of autosomal aneuploidies were estimated in surplus embryos that were further cultured to the blastocyst stage. The study design was unique, as embryo biopsy was performed twice—at the cleavage and the blastocyst stage—in the same embryo. In the present study, the total abnormality rate of the blastocysts was not significantly lower than at the cleavage stage. However, when we restricted our analysis to autosomal abnormalities, the abnormality rate of the blastocysts became significantly lower than at the cleavage stage. The rate of sex chromosome abnormality in the blastocysts was also significantly lower than at the cleavage stage. A reduction in aneuploidy from the cleavage to the blastocyst stage has been reported by others [22]. According to the available data, this reduction is due to the selection of normal cells in embryonic development [23]. However, the insignificant decrease in the abnormality rate of embryos that were examined at the blastocyst stage in the present study may be related to the small study population and differences in the techniques used.
Numerous studies have shown that different mechanisms are involved in the process of abnormality correction. These include multipolar division for trisomic rescue, fragment resorption, endoreduplication in cases of uniparental disomy for monosomic errors, blastomere exclusion via selection against highly aneuploid blastomeres to overcome chromosomal instability during the morula-blastocyst transition, encapsulation into micronuclei, elimination via cellular fragmentation, apoptosis of aneuploid cells, migration of euploid cells within the ICM, and migration of the aneuploid population to the blastocyst TE cells [1,2,4,24]. In addition, we reported that chaotic aneuploidy had a frequency of 13.23% in the blastocysts. One reason for chaotic aneuploidy is multipolar division [4]. Additionally, we reported a 14.70% rate of mosaicism in the blastocysts. The women in our study were 25–35 years old. However, Daughtry and Chavez [1] reported that mosaicism was prevalent in cases with advanced maternal age. Furthermore, we reported a 14.70% rate of sex chromosome abnormality as determined via FISH. In contrast, Liang et al. [25] reported a 6.5% rate of sex chromosome abnormality as determined using oligonucleotide DNA microarray analysis in blastocysts. Interestingly, we did not observe any abnormalities of chromosome 21, while Liang et al. [25] reported a 5% rate of chromosome 21 abnormality in blastocyst . Our findings showed that 29.4% of embryos at the cleavage stage exhibited aneuploidy of a sex chromosome versus 14.70% in the blastocyst stage. Zakharova et al. [14] declared that 44% of the embryos at the cleavage stage exhibited aneuploidy of a sex chromosome.
Kort et al. [23] also reported a lower rate of nuclear abnormalities in the blastocyst stage (5%) than in cleavage-stage embryos (16%), but the population of that study was larger than that of the present study. In our study, the total rate of abnormalities in the blastocyst stage was 32.35%. the genetic condition of blastocyst is more stable than the cleavage stage because genomic errors occur in different steps in oocytes, meiosis I and II [7], and fertilization, and about 50% of errors happen in post-fertilization mitotic division, causing mosaicism [26,27]. Furthermore, Lagalla et al. [2] reported that mitotic anomalies could become amended in the blastocyst stage. Barbash-Hazan et al. [22] also reported that some embryos have the potential to normalize their chromosomal status to proceed to the blastocyst stage [1]. Similar to previous studies, we found that the morphological characteristics of embryos did not exactly align with the chromosomal content, because 22 of 68 high-quality embryos had chromosomal abnormalities [15], whereas all of the aneuploid embryos had reached the blastocyst stage similar to the euploid embryos.
In this study, biopsy was performed at both the cleavage and blastocyst stages. Performing biopsy on ICM cells in addition to TE cells would facilitate a better understanding of the genetic correlation; however, that is considered an unethical procedure in a clinical setting. Munne and colleagues reported that the genetics of TE were different from ICM cells [28]. The main limitation of the FISH method is its capacity to analyze only a limited number of chromosomes [29,30]. Other flaws of FISH include signal overlap, hybridization failure, the lack of individual hybridization, the interpretation of results in the contiguous signals, the capacity to examine only one-third of the chromosomes, and the ambiguity of results for the other chromosomes [7]. Nowadays, newer technologies, such as comparative genomic hybridization microarray and next-generation sequencing, can be used to increase the accuracy of diagnosis of embryonic chromosomal abnormalities. However, these methods are not available in the majority of laboratories worldwide and are not cost-effective [31]. Therefore, the FISH method can be still considered a useful tool for identification of common chromosomal errors in embryos at both cleavage and blastocyst stages [31].
Another nuance that should be mentioned is that mosaic embryos may produce different results dependent on the population biopsied [2]. Therefore, we should be careful when making decisions about the presence of false or true mosaicism in blastocysts [1]. According to our results and previous reports, no doubt should exist that biopsy at the blastocyst stage is superior to biopsy at the cleavage stage. Sahin et al. [7] concluded that morula embryos had many abnormalities and that self-correction is disabled at this stage. Blastocyst biopsy facilitates the presence of a greater number of cells and a more informed genetic decision, such as in the contexts of mosaicism and chaotic aneuploidy in embryos [13]. Embryos can perform self-rescue by one of the mechanisms previously mentioned [1,2,4,24]. In the blastocyst, the embryo represents the first visible stage of differentiation, past the primary steps of cell differentiation. Therefore, embryos are known to have a notable implantation potential and a lower rate of aneuploidy compared to the cleavage stage [14,31]. Despite all of the mentioned advantages, less than 1% to 7% of biopsies are done in the blastocyst stage [16].
In conclusion, we recommend embryo biopsy at the TE stage rather than the cleavage stage in sex selection programs. The results of this study also emphasize that the examination of common chromosomal aneuploidies apart from sex selection cycles can be conducted in the blastocyst stage with the FISH method.
Acknowledgments
We would like to thank the staff of Assisted Reproductive Technologies at the Avicenna Infertility Center in Tehran and the staff of the Abortion Research Center at the Yazd Reproductive Sciences Institute in Yazd, Iran.
This study was supported by a grant from the Yazd Reproductive Sciences Institute (grant no: 5647).
Figure 1.
Flowchart of the chromosomal assessment of embryos in the cleavage and blastocyst stages via fluorescence in situ hybridization (FISH).
Figure 2.
The probes from the X series, which include green (chromosome 13), blue (18), red (21), and orange (X). The probes from the Y series include green (chromosome 15), blue (16), red (22), and orange (Y). (A) Trophectoderm (TE) cells from a hatching blastocyst, with approximately 40 cells biopsied at ×20 magnification. (B) Mosaicism of TE cells for the Y series at ×60 magnification. Green: chromosome 15 (2), blue: chromosome 16 (3), red: chromosome 22 (3), orange: Y chromosome (0), green: chromosome 15 (2), blue: chromosome 16 (2), red: chromosome 22 (2), orange: Y chromosome (0). (C) Triploid and chaotic TE cells with the probe Y series at ×60 magnification. Green: chromosome 15 (3), blue: chromosome 16 (3), red: chromosome 22 (3), orange: Y chromosome (0). (D) Normal cells for the X series from TE cells at ×60 magnification. Green: chromosome 13 (2), blue: chromosome 18 (2), red: chromosome 21 (2), orange: X chromosome (2). (E) X series for a euploid blastomere at ×100 magnification. One blastomere from an embryo in the cleavage stage. Green: chromosome 13 (2), blue: chromosome 18 (2), red: chromosome 21 (2), orange: X chromosome (2). (F) Y series for a euploid blastomere at ×100 magnification. Green: chromosome 15 (2), blue: chromosome 16 (2), red: chromosome 22 (2), orange: Y chromosome (2). (G) Tetraploid blastomere from the X series at ×100 magnification. Green: chromosome 13 (4), blue: chromosome 18 (4), red: chromosome 21 (4).
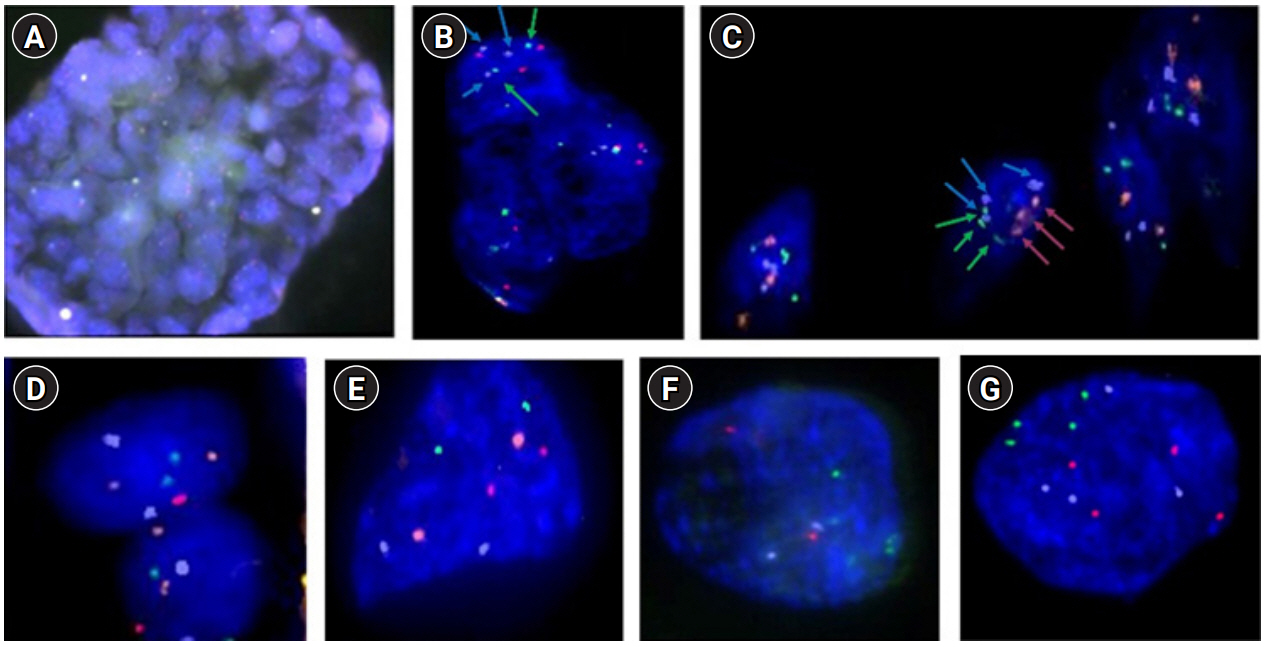
Table 1.
FISH results from two embryonic stages (cleavage and blastocyst)
| Chromosome | FISH (washing slides) in cleavage-stage embryos (n=68) | FISH in blastocysts (n=68) | p-value |
|---|---|---|---|
| 13 | 14 (20.4) | 0 | <0.001a) |
| 18 | 5 (22) | 1 (1.47) | 0.208a) |
| 21 | 14 (20.5) | 0 | <0.001a) |
| X | 19 (27.9) | 15 (22.05) | 0.428b) |
| 15 | 13 (19.1) | 8 (11.76) | 0.235b) |
| 16 | 18 (26.4) | 8 (11.76) | 0.029b) |
| 22 | 11 (16.1) | 7 (10.29) | 0.312b) |
| Y | 21 (30.8) | 6 (8.82) | 0.001b) |
| Autosomal abnormality | 30 (44.11) | 12 (17.64) | 0.008b) |
| Sex abnormality | 20 (29.4) | 10 (14.70) | 0.038b) |
| Total abnormal | 31 (45.58) | 22 (32.35) | 0.113b) |
| Total normal | 37 (54.41) | 46 (67.64) | 0.114b) |
| Chaotic | 9 (13.23) | ||
| Polyploid | 5 (7.35) | ||
| Mosaic | 10 (14.70) |
References
1. Daughtry BL, Chavez SL. Chromosomal instability in mammalian pre-implantation embryos: potential causes, detection methods, and clinical consequences. Cell Tissue Res 2016;363:201-25.



2. Lagalla C, Tarozzi N, Sciajno R, Wells D, Di Santo M, Nadalini M, et al. Embryos with morphokinetic abnormalities may develop into euploid blastocysts. Reprod Biomed Online 2017;34:137-46.


3. Gianaroli L, Magli MC, Ferraretti AP, Lappi M, Borghi E, Ermini B. Oocyte euploidy, pronuclear zygote morphology and embryo chromosomal complement. Hum Reprod 2007;22:241-9.



4. Daughtry BL, Rosenkrantz JL, Lazar NH, Fei SS, Redmayne N, Torkenczy KA, et al. Single-cell sequencing of primate preimplantation embryos reveals chromosome elimination via cellular fragmentation and blastomere exclusion. Genome Res 2019;29:367-82.



5. Scott RT Jr, Upham KM, Forman EJ, Hong KH, Scott KL, Taylor D, et al. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: a randomized controlled trial. Fertil Steril 2013;100:697-703.


6. Fragouli E, Bianchi V, Patrizio P, Obradors A, Huang Z, Borini A, et al. Transcriptomic profiling of human oocytes: association of meiotic aneuploidy and altered oocyte gene expression. Mol Hum Reprod 2010;16:570-82.



7. Sahin L, Bozkurt M, Sahin H, Gurel A, Yumru AE. Is preimplantation genetic diagnosis the ideal embryo selection method in aneuploidy screening? Kaohsiung J Med Sci 2014;30:491-8.


8. Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR, et al. The nature of aneuploidy with increasing age of the female partner: a review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil Steril 2014;101:656-63.


9. Munne S, Chen S, Colls P, Garrisi J, Zheng X, Cekleniak N, et al. Maternal age, morphology, development and chromosome abnormalities in over 6000 cleavage-stage embryos. Reprod Biomed Online 2007;14:628-34.


10. Lee E, Illingworth P, Wilton L, Chambers GM. The clinical effectiveness of preimplantation genetic diagnosis for aneuploidy in all 24 chromosomes (PGD-A): systematic review. Hum Reprod 2015;30:473-83.



11. Donoso P, Staessen C, Fauser BC, Devroey P. Current value of preimplantation genetic aneuploidy screening in IVF. Hum Reprod Update 2007;13:15-25.



12. Harton GL, Magli MC, Lundin K, Montag M, Lemmen J, Harper JC, et al. ESHRE PGD Consortium/Embryology Special Interest Group: best practice guidelines for polar body and embryo biopsy for preimplantation genetic diagnosis/screening (PGD/PGS). Hum Reprod 2011;26:41-6.



13. Sachdev NM, Maxwell SM, Besser AG, Grifo JA. Diagnosis and clinical management of embryonic mosaicism. Fertil Steril 2017;107:6-11.


14. Zakharova EE, Zaletova VV, Krivokharchenko AS. Biopsy of human morula-stage embryos: outcome of 215 IVF/ICSI cycles with PGS. PLoS One 2014;9:e106433.



15. Fesahat F, Kalantar SM, Sheikhha MH, Saeedi H, Montazeri F, Firouzabadi RD, et al. Developmental and cytogenetic assessments of preimplantation embryos derived from in-vivo or in-vitro matured human oocytes. Eur J Med Genet 2018;61:235-41.


16. De Rycke M, Goossens V, Kokkali G, Meijer-Hoogeveen M, Coonen E, Moutou C. ESHRE PGD Consortium data collection XIV-XV: cycles from January 2011 to December 2012 with pregnancy follow-up to October 2013. Hum Reprod 2017;32:1974-94.



17. Anbari F, Halvaei I, Nabi A, Ghazali S, Khalili MA, Johansson L. The quality of sperm preparation medium affects the motility, viability, and DNA integrity of human spermatozoa. J Hum Reprod Sci 2016;9:254-8.



18. World Health Organization. Sperm preparation techniques. In: World Health Organization, editor. WHO laboratory manual for the examination and processing of human semen. Geneva: World Health Organization; 2010. p. 160-68.
19. Sakkas D, Gardner DK. Evaluation of embryo quality analysis of morphology and physiology. In: Gardner DK, Weissman A, Howles CM, Shoham Z, editors. Textbook of assisted reproductive techniques. 5th ed. Boca Raton (FL): CRC Press; 2017. p. 226-43.
20. Magli MC, Jones GM, Lundin K, van den Abbeel E. Atlas of human embryology: from oocytes to preimplantation embryos. Preface. Hum Reprod 2012;27 Suppl 1:i1.
21. Singh S, Hobeika E, Knochenhauer ES, Traub ML. Pregnancy rates after pre-implantation genetic screening for aneuploidy are only superior when trophectoderm biopsy is performed on hatching embryos. J Assist Reprod Genet 2019;36:621-8.




22. Barbash-Hazan S, Frumkin T, Malcov M, Yaron Y, Cohen T, Azem F, et al. Preimplantation aneuploid embryos undergo self-correction in correlation with their developmental potential. Fertil Steril 2009;92:890-6.


23. Kort DH, Chia G, Treff NR, Tanaka AJ, Xing T, Vensand LB, et al. Human embryos commonly form abnormal nuclei during development: a mechanism of DNA damage, embryonic aneuploidy, and developmental arrest. Hum Reprod 2016;31:312-23.



24. Delhanty JD. The origins of genetic variation between individual human oocytes and embryos: implications for infertility. Hum Fertil (Camb) 2013;16:241-5.


25. Liang L, Wang CT, Sun X, Liu L, Li M, Witz C, et al. Identification of chromosomal errors in human preimplantation embryos with oligonucleotide DNA microarray. PLoS One 2013;8:e61838.



26. Fragouli E, Alfarawati S, Spath K, Jaroudi S, Sarasa J, Enciso M, et al. The origin and impact of embryonic aneuploidy. Hum Genet 2013;132:1001-13.



27. Capalbo A, Bono S, Spizzichino L, Biricik A, Baldi M, Colamaria S, et al. Sequential comprehensive chromosome analysis on polar bodies, blastomeres and trophoblast: insights into female meiotic errors and chromosomal segregation in the preimplantation window of embryo development. Hum Reprod 2013;28:509-18.



28. Munne S, Grifo J, Wells D. Mosaicism: "survival of the fittest" versus "no embryo left behind". Fertil Steril 2016;105:1146-9.


29. Zhang XY, Ata B, Son WY, Buckett WM, Tan SL, Ao A. Chromosome abnormality rates in human embryos obtained from in-vitro maturation and IVF treatment cycles. Reprod Biomed Online 2010;21:552-9.


- TOOLS







