Sperm DNA fragmentation and sex chromosome aneuploidy after swim-up versus density gradient centrifugation
Article information
Abstract
Objective
The aim of this study was to compare the efficacy of swim-up and density gradient centrifugation (DGC) for reducing the amount of sperm with fragmented DNA, sex chromosome aneuploidy, and abnormal chromatin structure.
Methods
Semen samples were obtained from 18 healthy male partners who attended infertility clinics for infertility investigations and were processed with swim-up and DGC. The percentages of sperm cells with fragmented DNA measured by the sperm chromatin dispersion test, normal sex chromosomes assessed by fluorescence in situ hybridization, and abnormal chromatin structure identified by toluidine blue staining were examined.
Results
The percentage of sperm cells with fragmented DNA was significantly lower in the swim-up fraction (9.7%, p=0.001) than in the unprocessed fraction (27.0%), but not in the DGC fraction (27.8%, p=0.098). The percentage of sperm cells with normal X or Y chromosomes was comparable in the three fractions. The percentage of sperm cells with abnormal chromatin structure significantly decreased after DGC (from 15.7% to 10.3%, p=0.002). The swim-up method also tended to reduce the percentage of sperm cells with abnormal chromatin structure, but the difference was not significant (from 15.7% to 11.6%, p=0.316).
Conclusion
The swim-up method is superior for enriching genetically competent sperm.
Introduction
The percentage of sperm cells with DNA fragmentation negatively influences natural and assisted conception [1] and is associated with high miscarriage rates in assisted conception [2]. Aneuploid sperm has been reported to be associated with the formation of aneuploid embryos, lower pregnancy and live birth rates, repeated intracytoplasmic sperm injection failure, and higher miscarriage rates [34]. Therefore, the selection of sperm without DNA fragmentation or aneuploidy is important for enhancing the outcomes of assisted conception.
Swim-up and density gradient centrifugation (DGC) are commonly used sperm preparation methods for assisted conception [5]. Their effects in terms of reducing the percentage of sperm DNA fragmentation (SDF) appears to be different based on the method used for assessing SDF and the study participants. Some studies reported that DGC preparation was superior to swim-up for reducing SDF levels [67], but others reported a similar efficacy [89]. Regarding sperm aneuploidy, studies have separately evaluated the efficacy of swimup [1011] and DGC [12] for lowering the frequency of aneuploidy.
However, no study has compared the two methods directly. Moreover, no studies have evaluated SDF levels and the sperm aneuploidy rate after the application of both methods at the same time on sperm samples from a single patient. In the present study, we compared the efficacy of the swim-up and DGC methods for reducing the SDF level, sex chromosome aneuploidy, and abnormal chromatin structure.
Methods
1. Study subjects
A comparative study was performed during 2015. Informed consent was obtained from all subjects, and the use of human semen for this study was approved by the Institutional Review Board of the Seoul National University Bundang Hospital (IRB No. B-1412-277-006). Semen samples were obtained from 18 healthy male partners who attended our infertility clinics for infertility investigations (a single sample per person). None of the subjects had a history of genital inflammation or surgery, no subjective symptoms or self-reported medical risk factors were identified, and none of the subjects had taken any prescribed medications. One subject was a smoker, and fertility was proven in seven subjects. The median age of the subjects was 35.5 years (range, 31–47 years).
2. Semen analysis
Semen samples were collected by masturbation after 3 days of sexual abstinence. After liquefaction for 30 minutes at room temperature, semen analysis was performed using motion analysis equipment (SAIS-PLUS 10.1; Medical Supply Co., Seoul, Korea), as described previously [13]. Strict criteria for the definition of normal sperm were used during the morphological assessment. In accordance with the World Health Organization guidelines (fifth edition, 2010), normal samples were defined by semen volume ≥1.5 mL, sperm count ≥15 million/mL, motility ≥40%, and a percentage of normal form of ≥4%. Round cells were counted (without peroxidase staining), and all the studied samples were found to contain <1 million/mL round cells. The basic semen characteristics were as follows (median [range]): semen volume, 3.0 mL (1.5–9.5 mL); sperm count, 75.5 million/mL (12.8–152 million/mL); total motility, 45.9% (25.1%–75%); total motile sperm count, 81 million (7.2–552 million); and percentage of normal form, 5.0% (1.5%–8.1%).
3. Sperm preparation
After a routine semen examination, the remainder of each semen sample was divided into three aliquots. One aliquot was processed by the swim-up method as previously described [13] (the swim-up fraction), and the other aliquot was processed by the DGC method (the DGC fraction). The third aliquot was not processed and served as the control (raw semen).
In the swim-up method, after initial centrifugation of the semen (300×g for 5 minutes) for removal of the seminal plasma, the pellet was suspended in fresh Ham's F10 medium (1.5 mL) supplemented with 10% serum substitute supplement (Irvine Scientific, Santa Ana, CA, USA) [13]. The mixture was again centrifuged (300×g for 5 minutes); the supernatant was discarded; and Ham's F10 media with 10% serum substitute supplement (0.5 mL) was gently layered on the pellet, followed by incubation at 37℃ in a 5% CO2 atmosphere for 1 hour. The supernatant (0.5 mL) was then transferred to a conical tube and later used for further experiments.
In the DGC method, the semen samples were processed by discontinuous silane-coated silica gradient (Sydney IVF Sperm Gradient; COOK Medical, Brisbane, Australia) as described in the kit instructions. After initial centrifugation of the semen sample (300×g for 5 minutes) for removal of the seminal plasma, the pellet was suspended in fresh Ham's F10 medium (3 mL) supplemented with 10% serum substitute supplement. The prewashed semen (3 mL) was then layered on top of a discontinuous silane-coated silica gradient in a 15-mL conical tube (40%/80%). Next, the conical tube was centrifuged at 300×g for 5 minutes, and the sperm collected from the bottom layer (80% layer) was washed twice by resuspension in 4 mL of Ham's F10 medium, followed by centrifugation (300×g for 5 minutes). After double centrifugation, the supernatant was removed and the pellet was resuspended in 1 mL of Ham's F10 medium supplemented with 10% serum substitute supplement. The resuspended pellet was later used for further experiments.
4. Measurements of SDF
SDF levels were measured using the Halosperm assay (Halotech DNA, Madrid, Spain) as described previously [1415]. Semen samples were diluted with Ham's F-10 medium (Sage, Trumbull, CT, USA) to adjust the concentration to 5 to 10 million/mL. Agarose gel from the kit was incubated for 5 minutes at 90℃ to 100℃ and then for 5 minutes at 37℃; this was followed by adding and mixing 25 µL of semen in a marked Eppendorf test tube. The mixtures (15 µL) were then placed on a super-coated slide and covered with a coverslip. The slides were kept for 5 minutes at 4℃ in a refrigerator to create a microgel with the implanted sperm. The coverslips were then carefully removed, followed by immersion of the slides into the prepared acid denaturation solution (80 µL in 10 mL of distilled water) for 7 minutes. The slides were then transferred to a tray with a lysis solution from the kit and incubated for 25 minutes. Next, the slides were rinsed with distilled water, followed by dehydration for 2 minutes in increasing concentrations of ethanol (70%, 90%, and 100%). After drying, the slides were stained with Diff-quick solution I and II (each for 7 minutes), rinsed with distilled water, and allowed to dry at room temperature.
Finally, each slide was examined under a light microscope at ×100 magnification, and at least 300 sperm cells were scored. Each sperm cell was categorized as having a large halo, medium halo, small halo, no halo, or degraded. A large halo meant that the sperm cell produced a halo with a thickness equal to or greater than the length of the minor diameter of the core, a medium halo meant that the sperm produced a halo with a thickness smaller than the length of the minor diameter of the core and greater than one-third of the minor diameter of the core, and a small halo meant that the sperm produced a halo with a thickness equal to or smaller than one-third of the minor diameter of the core. Sperm cells with a small halo or no halo, and those that were degraded, were classified as sperm cells with fragmented DNA.
5. Fluorescence in situ hybridization
Simultaneous two-probe two-color fluorescence in situ hybridization was performed using a probe set for the X and Y chromosomes. The probes used for fluorescence in situ hybridization were from loci DXZ1 and DYZ3. All the probes were directly labeled with fluorescent dyes; the X chromosome was labeled with Spectrum Orange, and the Y chromosome was labeled with Spectrum Green (Vysis Inc., Abbott Park, IL, USA). The detailed processes for the denaturation, hybridization, and 4’,6-diamidino-2-phenylindole (DAPI) counterstaining were performed as previously described [1617]. At least 300 sperm cells per slide were analyzed under a Nikon E600 epifluorescent microscope (Tokyo, Japan).
6. Toluidine blue staining
Chromatin structure was assessed by toluidine blue staining, as described previously [13]. Based on a visual assessment, the stained sperms were categorized as having the following toluidine blue colors: dark purple, intermediate, and light blue. Only dark purple sperm cells were considered to have an abnormal chromatin structure. At least 300 sperm cells were counted per slide, and the percentage of sperm cells with a dark-purple color was determined.
7. Statistical analysis
Statistical analysis was performed using MedCalc ver. 10.4 (MedCalc Software, Mariakerke, Belgium). The SDF level, the percentage of normal X or Y sperm cells, and the percentage of sperm cells with an abnormal chromatin structure were compared between the raw and processed semen samples using the Wilcoxon signed-rank test. The Spearman correlation test was used to assess associations between different semen parameters. A p-value of <0.05 was considered to indicate statistical significance.
Results
The SDF level in the swim-up fraction (9.7%) was significantly lower than in the unprocessed fraction (27.0%, p=0.001), but not in the DGC fraction (27.8%, p=0.098) (Table 1). The SDF level in the swim-up fraction was also significantly lower than in the DGC fraction (p=0.001). The percentage of normal X or Y sperm was similar in the three fractions. The percentage of sperm cells with abnormal chromatin structure was significantly lower in the DGC fraction than in the unprocessed fraction (10.3% vs. 15.7%, p=0.002), but not in the swim-up fraction (11.6%, p=0.316).

Percentages of sperm cells with fragmented DNA, normal X or Y sperm cells, and sperm cells with abnormal chromatin structure after SU versus DGC
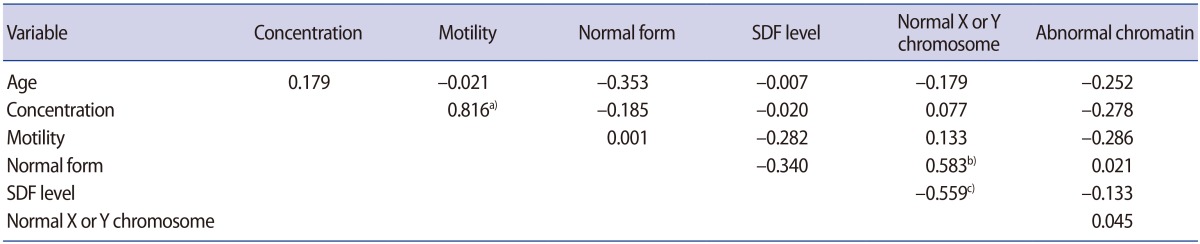
The correlation coefficients between various parameters in the raw semen are listed in Table 2. A statistically significant positive correlation was observed between the percentage of normal X or Y sperm and the percentage of normal form (r=0.583, p=0.007). A statistically significant negative correlation was observed between the percentage of normal X or Y sperm and the SDF level (r=−0.559, p=0.042). The percentage of normal form showed no associations with the SDF level. The percentage of sperm with abnormal chromatin structure had no correlation with any of the parameters listed.
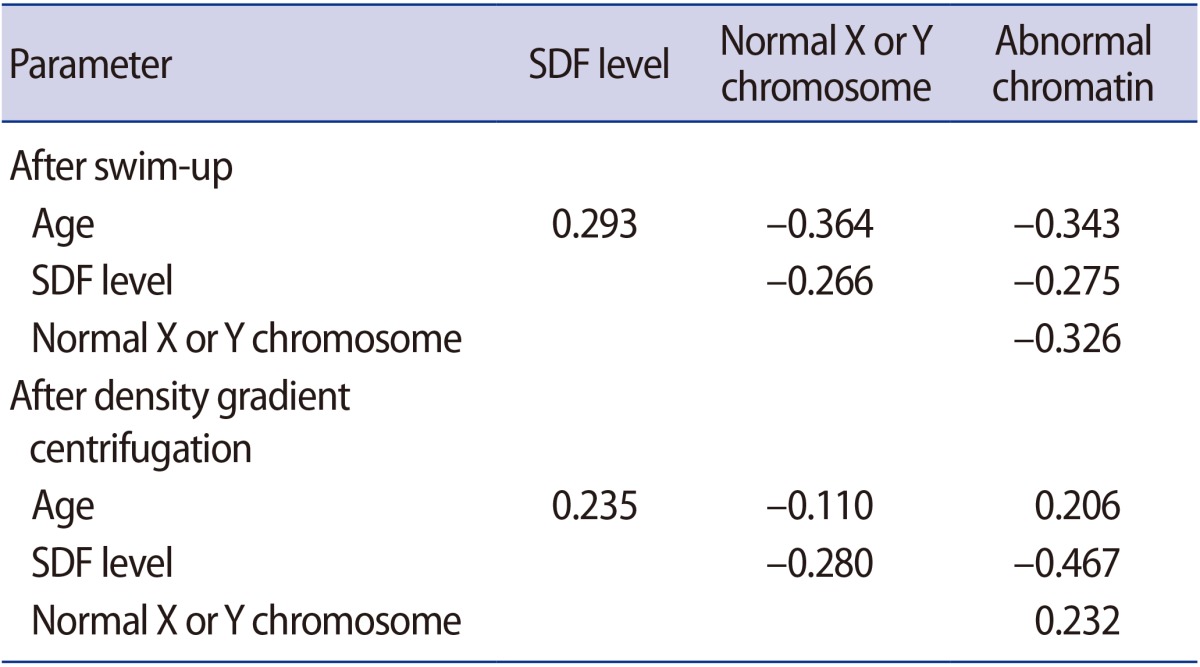
In the processed semen, no associations were found between the percentage of normal X or Y sperm, the percentage of sperm with abnormal chromatin structure, and the SDF level (Table 3).
Discussion
In the present study, swim-up was found to be more effective than DGC for reducing the amount of sperm with fragmented DNA. DGC was more effective for reducing the amount of sperm with abnormal chromatin structure, but the swim-up method also tended to do so, even if it was statistically not significant. Both methods had no effect on the preservation of normal X or Y sperm. Based on these findings, swim-up is better than DGC for enriching genetically competent sperm.
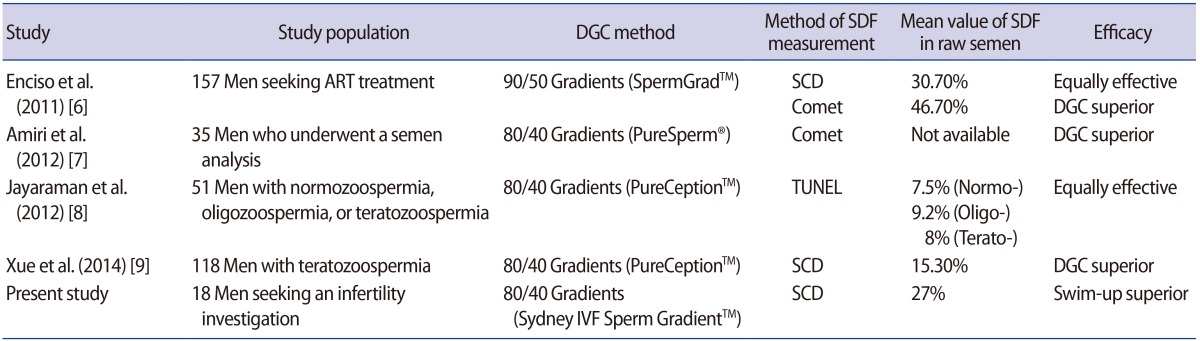
In previous studies, the populations and the methods used for DGC or the measurement of SDF were heterogeneous (Table 4) [6789]. Two studies were similar to our study in terms of the study population. They found DGC to be superior to the swim-up method for reducing SDF levels, as measured using the Comet assay [67]. However, Enciso et al. [6] found a similar efficacy of both preparation methods for reducing SDF levels measured using the sperm chromatin dispersion (SCD) test; the mean SDF levels significantly decreased in both the swim-up (mean, 16%) and the DGC (mean, 19.8%) compared with those of neat semen.
In the present study, the mean levels of SDF significantly decreased after swim-up, but not after DGC. A possible reason for the ineffectiveness of DGC in reducing SDF levels is that our DGC protocol, which contains a number of centrifugations, might have had a deleterious effect on sperm DNA integrity. Centrifugation generates radical oxygen species, which have been reported to be a mechanism causing SDF [18].
The Halosperm assay was used to measure the SDF level. This test is easy and yields rapid results [19]. Furthermore, it is relatively inexpensive because it does not require immunofluorescence equipment. SDF levels measured by the SCD test are typically higher than those measured using the terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick-end labeling (TUNEL) method because SCD has been suggested to have greater sensitivity than the TUNEL method [20]. However, one study reported similar SDF levels using these two methods [21].
The DGC was effective at reducing the amount of sperm with abnormal chromatin structure in this study. Swim-up showed a modest effect, although it was not statistically significant. Marchesi et al. [22] also found that both methods had a similar efficacy for reducing the amount of sperm with abnormal chromatin structure. In general, abnormal chromatin structure is an indicator of sperm maturity. Kovanci et al. [12] observed a positive relationship between the disomy rate and sperm immaturity, which suggested that disomy primarily originates in immature sperm. However, we did not find any association between the percentage of normal X or Y sperm and the percentage of sperm with abnormal chromatin structure.
Neither sperm preparation method had any effect on the percentage of normal X or Y sperm because of the high percentage of sperm with normal sex chromosomes in the raw semen of our study population, who had normal sperm or mild sperm abnormalities. Ong et al. [10] also reported that the sex chromosome disomy rate did not decrease after swim-up in 20 healthy donors. However, a significant decrease in sex chromosome disomy was reported after swim-up [11] or DGC [1223] in several studies including men with subnormal or abnormal sperm. Both methods reduced aneuploidy in men with subnormal or abnormal sperm, whose sex chromosome aneuploidy rate is higher [24].
In the raw semen, the percentage of normal X or Y sperm showed a positive association with the percentage of normal form and a negative association with the SDF level. This indicated that sperm cells with normal sex chromosomes tended to have a normal shape and less DNA fragmentation. To date, no studies have investigated the relationships among these three sperm parameters. Sperm aneuploidy has been hypothesized to trigger SDF via an apoptosis-like process mediated by endogenous nucleases [25]. It is suggested that same mechanisms might be involved in sperm DNA damage and aneuploidy formation [2627].
Interestingly, no relationship was found between the percentage of normal X or Y sperm and the SDF level after sperm processing using either swim-up or DGC. This may have been due to the increase in sperm quality caused by the sperm processing. In our previous work, sperm motility and the percentage of normal form increased and the SDF level decreased significantly after swim-up [13].
Although swim-up efficiently removes sperm with fragmented DNA, approximately 10% of the sperm cells still had fragmented DNA. For intracytoplasmic sperm injection, the probability of selecting a sperm cell with fragmented DNA may be close to 10% even after swim-up. New technologies to select a single functionally competent sperm cell without genetic abnormalities need to be developed.
To the best of our knowledge, this is the first study evaluating SDF and sperm aneuploidy after the application of both methods at the same time on sperm samples from the same patient. This is also the first study to identify direct associations among normal form, the SDF level, and sex chromosome aneuploidy in raw semen. However, our study has some limitations. Because we analyzed semen samples from healthy male partners, the results of this study cannot be extrapolated to patients with severe male factor infertility.
In summary, processing sperm with swim-up is better than using DGC for enriching genetically competent sperm in infertile couples who do not have male factor infertility. Even if the percentage is decreased substantially, sperm cells with genetic abnormalities still remain after swim-up. It is expected that developing novel methods to conventional swim-up will enable the selection of functionally and genetically competent sperm cells one-by-one, just as in natural selection.
Acknowledgments
The authors wish to thank the staff of the Laboratory of Reproductive Medicine, Seoul National University Bundang Hospital, Seongnam, Korea.
Notes
This work was supported by the Korea Health Care Technology R&D Project, Ministry of Health and Welfare, Korea (No. A120043).
Conflict of interest: No potential conflict of interest relevant to this article was reported.