MTHFR 3′-untranslated region polymorphisms contribute to recurrent pregnancy loss risk and alterations in peripheral natural killer cell proportions
Article information
Abstract
Objective
To identify the associations between polymorphisms of the 3′-untranslated region (UTR) of methylenetetrahydrofolate reductase (MTHFR) gene, which codes for an important regulatory enzyme primarily involved in folate metabolism, and idiopathic recurrent pregnancy loss (RPL) in Korean women.
Methods
The study population comprised 369 RPL patients and 228 controls. MTHFR 2572C>A, 4869C>G, 5488C>T, and 6685T>C 3′-UTR polymorphisms were genotyped by polymerase chain reaction-restriction fragment length polymorphism analysis or by TaqMan allelic discrimination assays. Natural killer cell proportions were determined by flow cytometry.
Results
The MTHFR 2572-5488-6685 (A-C-T) haplotype had an adjusted odds ratio of 0.420 (95% confidence interval, 0.178–0.994; p=0.048) for RPL. Analysis of variance revealed that MTHFR 4869C>G was associated with altered CD56+ natural killer cell percentages (CC, 17.91%±8.04%; CG, 12.67%±4.64%; p=0.024) and folate levels (CC, 12.01±7.18 mg/mL; CG, 22.15±26.25 mg/mL; p=0.006).
Conclusion
Variants in the 3′-UTR of MTHFR are potential biomarkers for RPL. However, these results should be validated in additional studies of ethnically diverse groups of patients.
Introduction
One in eight pregnant women will experience the loss of a pregnancy, most frequently within 2 to 3 months of conception [1]. The possibility of an additional pregnancy loss is 5% higher for women who experienced the loss of their first pregnancy [2]. There are many etiologic factors that contribute to recurrent pregnancy loss (RPL), defined by the American Society for Reproductive Medicine as the consecutive loss of two or more pregnancies [34], including genetics, anatomical deformities, thrombophilia, endocrine dysfunction, placental anomalies, infection, smoking, excessive alcohol consumption, environmental factors, psychological trauma, and stress [5]. Recent studies investigating the pathophysiology of RPL have reported that a high level of homocysteine (Hcy) and a low level of folate in the blood might be a marker of RPL risk [678]
High levels of Hcy and/or a deficiency of folate in the plasma have been associated with reduced expression of the enzyme methylenetetrahydrofolate reductase (MTHFR), which catalyzes the conversion of 5, 10-methylenetetrahydrofolate to 5-methyltetrahydrofolate [9101112]. MTHFR is also a critical factor in DNA methylation and is an important regulatory enzyme primarily involved in folate metabolism [1314]. A number of studies have examined the association of RPL incidence with MTHFR polymorphisms, such as 677C>T (rs1801133) and 1298A>C (rs1801131) [1516]. The MTHFR 677T allele was closely correlated with decreased MTHFR activity, resulting in folate deficiency and increased plasma Hcy levels [11]. Furthermore, the 677T allele was determined to be a potential genetic risk factor for increased RPL susceptibility in recent meta-analyses [17]. Although an association between RPL and genetic variation of the 3′-untranslated region (UTR) of MTHFR has been reported [18], such variants and their associations with RPL have not been studied in detail [19]. Variations in the 3′-UTR may result in the inhibition of translation or degradation of mRNA [20], thereby altering the expression of the encoded protein. This study examined the 2572C>A (rs4846049), 4869C>G (rs1537514), 5488C>T (rs3737967), and 6685T>C (rs4846048) polymorphisms of MTHFR in pregnant women and their association with RPL.
Studies of patients with idiopathic RPL and in vitro fertilization-embryo transfer failure have shown that the numbers of decidual natural killer (NK) cells are significantly reduced in those patients [2122], indicating their importance during pregnancy [23]. In particular, the accumulation of CD56+ subsets of NK cells has been shown to be important during early pregnancy [2425]. As such, we also examined the numbers of NK cells and the proportions of CD56+ subtypes in pregnant women and those with RPL in relation to their MTHFR genotype.
Methods
1. Study population
For this study, 369 women with RPL and 228 controls were recruited from the CHA Bundang Medical Center. Written consent was obtained from all participants. The women included in the control group had a history of at least 1 successful naturally conceived pregnancy, no history of pregnancy loss, and a karyotype of 46,XX. The use of participants in this study was approved under institutional review board (No. BD2010-123D).
RPL patients were defined as those who had experienced at least two consecutive spontaneous abortions identified by ultrasonography, human chorionic gonadotropin levels, and/or physical examinations. Patients with a history of smoking or alcohol use were excluded from the study. RPLs resulting from anatomic abnormalities, such as intrauterine adhesions, septate uteri, and uterine fibroids, were determined by hysterosalpingography, hysteroscopy, computed tomography, and magnetic resonance imaging. Hormonal causes of miscarriages included hyperprolactinemia, luteal insufficiency, and thyroid disease, and were determined from the blood levels of the appropriate hormones. To verify a chromosomal cause for miscarriage, chromosome analysis was performed using standard protocols [26], and metaphase chromosomes were examined using the trypsin-Giemsa banding (GTG-banding) method. Bacterial cultures for Ureaplasma urealyticum and/or Mycoplasma hominis were used to identify miscarriages caused by infection. Autoimmune causes of RPL, defined as antiphospholipid syndrome and lupus, were evaluated using lupus anticoagulant and anticardiolipin antibodies. Thrombophilia, determined by deficiencies of protein C and protein S and the presence of anti-β-2 glycoprotein antibodies, was also considered as a cause of RPL.
2. Genotyping
Genomic DNA samples from RPL patients and controls were extracted from anticoagulated peripheral blood using the G-DEX blood extraction kit (Intron, Seongnam, Korea). MTHFR 2572C>A and MTHFR 4869C>G polymorphisms were genotyped by polymerase chain reaction-restriction fragment length polymorphism analysis using the following primers: 5′-TTG CCA ACT AAG CCC TCG AAA CAA-3′ (sense) and 5′-TGC CAC ATC TCT TCT ACG ATG CCA-3′ (antisense) for MTHFR 2572C>A, and 5′-TCC AGC CCT GAG CCC AGA GTC T-3′ (sense) and 5′-AGG CAA GCC CCT CAG CCCTT-3′ (antisense) for MTHFR 4869C>G. Restriction enzyme digestions, using StyI and BsmAI (New England BioLabs, Essex County, MA, USA), were performed for the MTHFR 2572C>A and MTHFR 4869C>G polymorphisms, respectively, at 37℃ for 16 hours.
MTHFR 5488C>T and MTHFR 6685T>C polymorphisms were assessed using real-time polymerase chain reaction (RG-6000; Corbett Research, Mortlake, Australia) with the following primers and probes: 5′-GAG GCA CCA GCT CTG TGG-3′ (forward) and 5′-CCC CAG GAA GTC CAA GC-3′ (reverse) with 5′-FAM-CAG CAG CTG CGG GTC TGA A-TAMRA-3′ (C allele) and 5′ JOE-CAG CAG CTG TGG GTC TGA A-TAMRA-3′ (T allele) for MTHFR 5488C>T, and 5′-CCA GAC CAG AAG CAG TTA-3′ (forward) and 5′-GCT GTG CAG TGT CAT TT-3′ (reverse) with 5′-FAMCAC CAA CAA ATG GTG ATA AG-TAMRA-3′ (T allele) and 5′-JOE-CAC CAA CAA GTG GTG ATA AG-TAMRA-3′ (C allele) for MTHFR 6685T>C.
3. Estimations of clinical characteristics from blood samples
Levels of Hcy, folate, total cholesterol, and uric acid were measured in plasma samples collected from RPL patients after 12 hours of fasting. Hcy was measured using a fluorescence polarization immunoassay with an Abbott IMx analyzer (Abbott Laboratories, Abbott Park, IL, USA). The total cholesterol levels and uric acid concentrations were estimated using commercially available enzymatic colorimetric tests by the Roche/Hitachi Modular Pre-analytics Plus system (Roche Diagnostics, Mannheim, Germany). Additionally, platelet counts, folate and plasminogen activator inhibitor-1 levels, prothrombin time, and activated partial thromboplastin time (aPTT) were determined.
4. Estimation of peripheral CD56+ NK cell proportions
Measurements of NK cells were performed by flow cytometry with CellQuest software (BD FACSCalibur; BD Biosciences, Seoul, Korea). Fluorescently labeled (fluorescein isothiocyanate, phycoerythrin [PE], peridinin chlorophyll protein, and allophycocyanin) monoclonal antibodies specific for CD3, CD16, and CD56 were purchased from BD Biosciences. Anti-NKG2A-PE antibodies were obtained from Immunotech (Beckman Coulter, Fullerton, CA, USA). Peripheral blood mononuclear cells (2.5×105) were stained for cell-surface antigen expression at 4℃ in the dark for 30 minutes, washed twice in 2 mL of phosphate-buffered saline containing 1% bovine serum albumin and 0.01% sodium azide (FACS wash buffer), and subsequently fixed in 200 µL of a 1% formaldehyde solution (Sigma-Aldrich, St. Louis, MO, USA) prior to sorting, as previously described [2728].
5. Statistical analysis
The associations between MTHFR polymorphisms and RPL risk were examined by odds ratios (ORs), adjusted odds ratios (AORs), and 95% confidence intervals (CIs). The data are presented as the mean numbers and percentages with standard deviations (categorical variables). Differences in the frequencies of MTHFR polymorphisms between the control and patient groups were assessed using the Fisher exact test and a logistic regression model. Statistical significance was accepted at p<0.05. The correlations of each genotype or allele with the proportion of NK cells and plasma Hcy, folate, total cholesterol, and uric acid levels were assessed by the Kruskal-Wallis and Mann-Whitney U tests. Statistical analyses were performed using GraphPad Prism 4.0 (GraphPad Software Inc., San Diego, CA, USA), MedCalc version 12.1.4 (MedCalc Software, Mariakerke, Belgium), and StatsDirect V2.6.6 (StatsDirect Ltd., Warrington, UK). Haplotype analysis was performed using SNPAlyze ver. 5.1 (Dynacom Co., Yokohama, Japan), Hapstat 3.0 (http://dlin.web.unc.edu/software/hapstat), and Haploview (https://www.broadinstitute.org/haploview/haploview).
Results
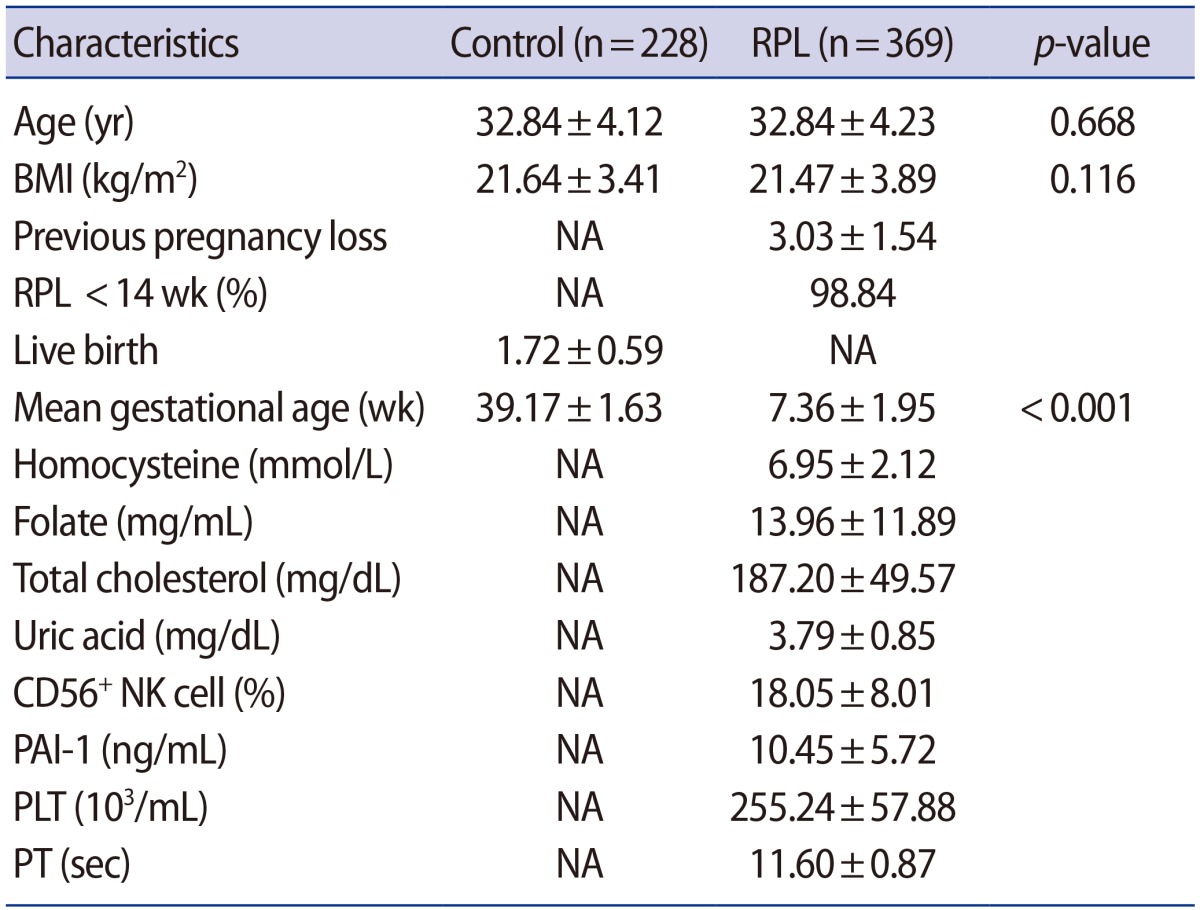
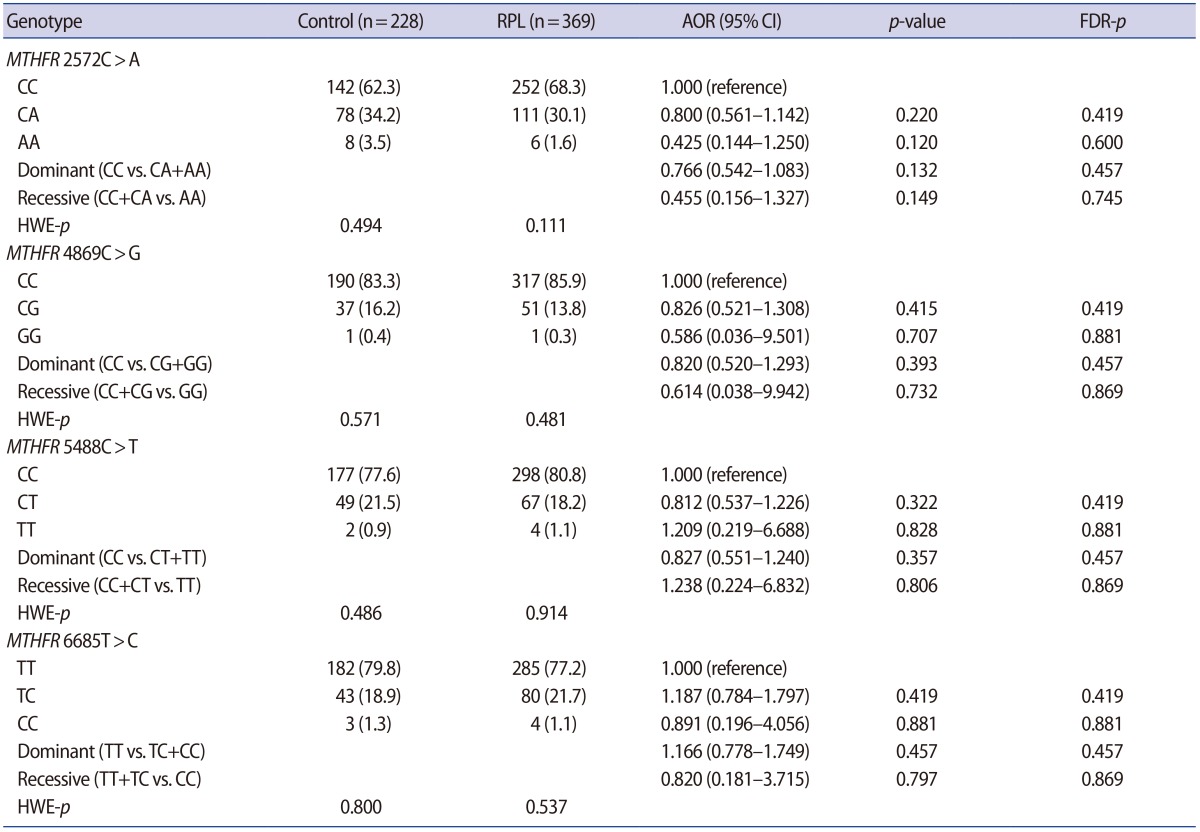
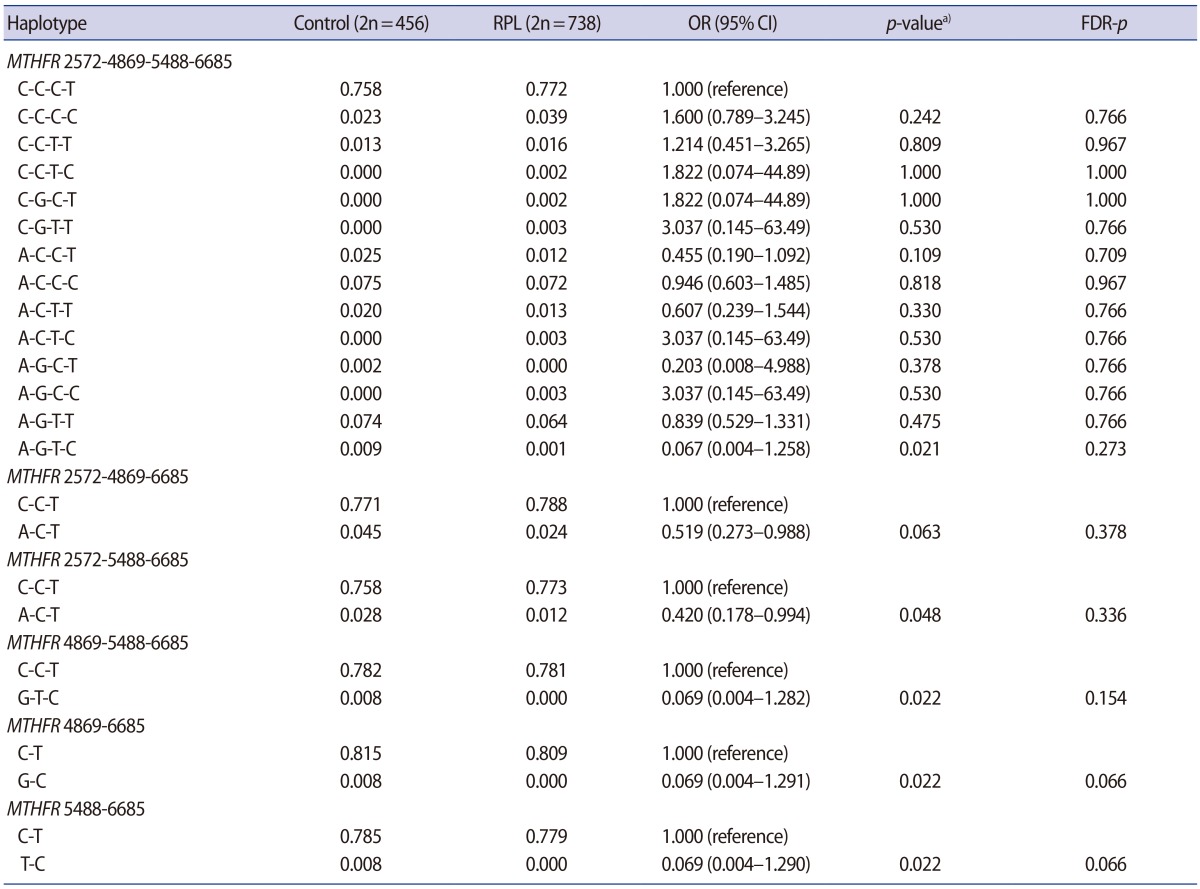
The clinical characteristics of the RPL patients and control subjects are presented in Table 1. The MTHFR genotype frequencies for controls and RPL patients were consistent with expectations under the assumption of Hardy-Weinberg equilibrium (Table 2). We further stratified the MTHFR polymorphism data among RPL patients with three or more pregnancy losses and those with four or more (Supplement 1). The genotypes were not associated with RPL. However, haplotype analyses for multiple MTHFR 3′-UTR polymorphisms revealed a negative correlation between RPL and the MTHFR 2572/5488/6685 model, in particular, with the A-C-T alleles (p=0.048) (Table 3). In addition, we conducted a haplotype analysis based on our previous studies to investigate the association of the well-known MTHFR 677C>T/1298A>C mutations with MTHFR 3′-UTR polymorphisms (Supplement 2), which showed that RPL was negatively correlated with the MTHFR 677-1298-1793-2572-4869-5488-6685 model, particularly, with C-C-G-A-C-C-T (OR, 0.026; 95% CI, 0.002–0.446; p<0.001). A significant association was also found with MTHFR 257 2CC/6685CT (AOR, 2.867; 95% CI, 1.068–7.696; p=0.037) in a combination analysis (Supplement 3). However, there were no associations with RPL or other genotypic combinations.
We conducted variance analyses for differences in clinical factors and genotypes in RPL patients (Figure 1, Supplements 4 and 5). We investigated the CD56+ NK cell proportions and plasma levels of Hcy, folate, total cholesterol, and uric acid, as well as blood coagulation factors such as prothrombin time, plasminogen activator inhibitor-1, platelet count, and aPTT. The percentages of CD56+ NK cells and folate levels were significantly different between the alleles of MTHFR 4869 in RPL patients with three or more lost pregnancies (p=0.024 and p=0.006, respectively) (Figure 1, Supplement 5), with a similar trend observed in women with the MTHFR 4869CG genotype and three or more lost pregnancies (Supplement 4).

The MTHFR 4869C>G polymorphism is associated with differences in CD56+ NK cell percentages and folate levels in women with recurrent pregnancy loss. Statistical analyses were performed using the Kruskal-Wallis test for the MTHFR 4869C>G genotype and the percentages of CD56+ NK cells (A) and folate levels (B) in women with three or more lost pregnancies. NK, natural killer; MTHFR, methylenetetrahydrofolate reductase. a)p<0.05.
The MTHFR 4869C>G polymorphism was also associated with plasma Hcy and folate levels (Supplement 6) and with CD56+ NK cell proportions and aPTT (Supplement 7) after adjusting for the ages of the participants. The age-adjusted analyses also revealed associations between the MTHFR 5488C>T dominant genotype and plasma Hcy, folate levels, and aPTT, as well as associations between the MTHFR 6685T>C dominant genotype and plasminogen activator inhibitor-1 levels and aPTT (Supplements 6, 7).
Discussion
In this study, we investigated four 3′-UTR polymorphisms of the MTHFR gene in relation to the occurrence of RPL. We found that the 2572C>A, 4869C>G, 5488C>T, and 6685T>C MTHFR genotypes were substantially associated with RPL susceptibility and displayed significant haplotype and environmental factor effects (occupational exposure, air pollution, etc.). Specifically, MTHFR 4869C>G and 5488C>T polymorphisms were associated with increased Hcy levels and decreased folate levels in plasma. To our knowledge, this is the first study to provide evidence linking 3′-UTR polymorphisms of MTHFR with RPL. Combined with previous findings linking MTHFR polymorphisms with increased RPL susceptibility [151617], our data implicate genetic variation in the 3′-UTR, further supporting the role of MTHFR in RPL.
Previous studies have reported haplotype analysis of MTHFR 677C>T and 1298A>C [61112141617], but the single nucleotide polymorphisms (SNPs) present in the 3′-UTR have not been analyzed. These additional SNPs may alter MTHFR expression or function, as was found with MTHFR 677C>T SNPs with regard to RPL. Moreover, newly identified SNPs may be useful as biomarkers. For these reasons, we conducted a study to examine the SNP genotypes and haplotypes of the 3′-UTR region, which is the microRNA (miRNA) binding site. We found that RPL was associated with higher frequencies of the 4869G and 5488T MTHFR alleles. These alleles were, in turn, associated with differences in Hcy and folate levels between controls and women with RPL. Furthermore, the MTHFR 4869G allele was associated with lower percentages of CD56+ NK cells, which has been linked with a favorable pregnancy result in women with RPL [29]. The activation of NK cells is an important stage in the inflammatory response and is known as a prognostic factor in women with RPL [30]. Interestingly, the frequency of the MTHFR 4869C>G polymorphism tended to decrease with an increasing number of lost pregnancies. Therefore, our study demonstrated a negative correlation between the MTHFR 4869G allele frequency and CD56+ NK cell proportion, indicating that the presence of this allele can impact the activity of CD+ NK cells.
In a previous study, the proportion of NK cells, depending on the MTHFR 677C>T polymorphism, was associated with an increased risk of RPL [16]. Similarly, we found that the proportion of NK cells was lower with MTHFR 4869CG than with MTHFR 4869CC. Therefore, the decrease in NK cells, which influence immune balance during pregnancy, was associated with RPL occurrence. Our results also indicated that the MTHFR 4869C>G polymorphism may be somewhat protective against RPL, although the difference was not statistically significant. However, the haplotypes with the MTHFR 4869G allele were significantly associated with a lower risk of RPL occurrence. Furthermore, we also found that the frequency of the MTHFR 2572A allele was lower in women with higher numbers of lost pregnancies, indicating that Korean women with this polymorphism are more susceptible to RPL.
Although the functional impacts of these genetic variations in the 3′-UTR region are not fully understood, these polymorphisms may alter miRNA binding affinities and affect the translation or stability of the MTHFR transcript [19]. For example, the MTHFR 2572C>A variant is associated with increased binding of miR-149 and reduced MTHFR expression [31]. Further studies of the effects of the other MTHFR 3′-UTR polymorphisms are needed to determine their effect on miRNA binding and MTHFR expression.
Several limitations of this study should be noted. First, this study did not control for additional environmental risk factors for RPL that may have influenced the results. Second, the study population was limited to Koreans, and our findings may therefore not be applicable to other populations. Lastly, the number of patients included in the subgroup analysis was relatively small.
In conclusion, this study examined four polymorphisms of the 3′-UTR of MTHFR in association with RPL prevalence in Korean women. We found that the MTHFR 4869C>G polymorphism was associated with lower percentages of CD56+ NK cells and higher folate levels in women with RPL. Therefore, variants in the 3′-UTR of MTHFR in women may represent potential biomarkers of RPL. However, additional studies of ethnically diverse groups of patients are needed to validate these findings.
Notes
This work was partly supported by a grant of the Korea Healthcare Technology R&D Project (HI15C1972010015), Ministry for Health, Welfare & Family Affairs, Republic of Korea and by Basic Science Research Program through National Research Foundation of Korea Grants (2009-0093821, 2017R1D1A1B03031542 and 2015R1D1A1A09057432) funded by the Korean Government, Korea.
Conflict of interest: No potential conflict of interest relevant to this article was reported.
References
Supplementary Materials
Supplement 1. Frequencies of MTHFR gene polymorphisms according to the number of pregnancy losses. Supplemental data can be found at: http://ecerm.org/src/sm/cerm-44-152-s001.pdf.
Supplement 2. Haplotype analysis of MTHFR gene polymorphisms in control subjects and RPL patients. Supplemental data can be found at: http://ecerm.org/src/sm/cerm-44-152-s002.pdf.
Supplement 3. Combined analysis of MTHFR gene polymorphisms in control subjects and RPL patients. Supplemental data can be found at: http://ecerm.org/src/sm/cerm-44-152-s003.pdf.
Supplement 4. Clinical parameters according to MTHFR gene polymorphisms for patients with two or more lost pregnancies. Supplemental data can be found at: http://ecerm.org/src/sm/cerm-44-152-s004.pdf.
Supplement 5. Clinical parameters according to MTHFR gene polymorphisms for patients with three or more lost pregnancies. Supplemental data can be found at: http://ecerm.org/src/sm/cerm-44-152-s005.pdf.
Supplement 6. Genetic associations between MTHFR gene polymorphisms and homocysteine and folate with adjustment for age. Supplemental data can be found at: http://ecerm.org/src/sm/cerm-44-152-s006.pdf.
Supplement 7. Associations of MTHFR gene polymorphisms with CD56+ NK cells, PAI-1, and aPTT with adjustment for age. Supplemental data can be found at: http://ecerm.org/src/sm/cerm-44-152-s007.pdf.
Supplement 1
Frequencies of MTHFR gene polymorphisms according to the number of pregnancy losses
Supplement 2
Haplotype analysis of MTHFR gene polymorphisms in control subjects and RPL patients
Supplement 3
Combined analysis of MTHFR gene polymorphisms in control subjects and RPL patients
Supplement 4
Clinical parameters according to MTHFR gene polymorphisms for patients with two or more lost pregnancies
Supplement 5
Clinical parameters according to MTHFR gene polymorphisms for patients with three or more lost pregnancies
Supplement 6
Genetic associations between MTHFR gene polymorphisms and homocysteine and folate with adjustment for age
Supplement 7
Associations of MTHFR gene polymorphisms with CD56+ NK cells, PAI-1, and aPTT with adjustment for age