Polymorphisms and expression levels of TNP2, SYCP3, and AZFa genes in patients with azoospermia
Article information
Abstract
Objective
Azoospermia (the total absence of sperm in the ejaculate) affects approximately 10% of infertile males. Despite diagnostic advances, azoospermia remains the most challenging issue associated with infertility treatment. Our study evaluated transition nuclear protein 2 (TNP2) and synaptonemal complex protein 3 (SYCP3) polymorphisms, azoospermia factor a (AZFa) microdeletion, and gene expression levels in 100 patients with azoospermia.
Methods
We investigated a TNP2 single-nucleotide polymorphism through polymerase chain reaction (PCR) restriction fragment length polymorphism analysis using a particular endonuclease. An allele-specific PCR assay for SYCP3 was performed utilizing two forward primers and a common reverse primer in two PCR reactions. Based on the European Academy of Andrology guidelines, AZFa microdeletions were evaluated by multiplex PCR. TNP2, SYCP3, and the AZFa region main gene (DEAD-box helicase 3 and Y-linked [DDX3Y]) expression levels were assessed via quantitative PCR, and receiver operating characteristic curve analysis was used to determine the diagnostic capability of these genes.
Results
The TNP2 genotyping and allelic frequency in infertile males did not differ significantly from fertile volunteers. In participants with azoospermia, the allelic frequency of the SYCP3 mutant allele (C allele) was significantly altered. Deletion of sY84 and sY86 was discovered in patients with azoospermia and oligozoospermia. Moreover, SYCP3 and DDX3Y showed decreased expression levels in the azoospermia group, and they exhibited potential as biomarkers for diagnosing azoospermia (area under the curve, 0.722 and 0.720, respectively).
Conclusions
These results suggest that reduced SYCP3 and DDX3Y mRNA expression profiles in testicular tissue are associated with a higher likelihood of retrieving spermatozoa in individuals with azoospermia. The homozygous genotype TT of the SYCP3 polymorphism was significantly associated with azoospermia.
Introduction
It is a public health concern that 7% of males experience fertility issues at some point in their lives. Male infertility is a complicated condition that, by international consensus, is defined by the failure to achieve a clinical pregnancy following 12 months of regular, unprotected sexual activity. Male factor infertility is present in 45% to 50% of couples affected by infertility [1].
A lack of sperm in the ejaculate is known as azoospermia [2]. Azoospermia always results in infertility, although the term itself does not describe the underlying causation. Global estimates indicate that up to 10% of infertile men and one out of every 100 males of reproductive age have azoospermia [3]. The two broad categories are obstructive azoospermia (OA) and nonobstructive azoospermia (NOA) [4]. This distinction has clinical significance because it impacts patient care and recovery [3]. Notably, NOA refers to an inherent testicular abnormality induced by several diseases that can significantly impact sperm production [5]. Primary testicular failure affecting spermatogenic cells is known as spermatogenic failure, and hypogonadotropic hypogonadism (HH) is a condition that results in a severe spermatogenic deficiency in patients with NOA [6].
OA, in contrast, is caused by a mechanical obstruction along the reproductive tract, specifically the vas deferens, epididymis, or ejaculatory duct [7]. Unlike NOA, spermatogenesis is preserved in patients with OA, and reconstructive operations and sperm retrieval are frequently very successful [2]. Abnormalities in sex chromosomes and a growing number of uncommon detrimental variations in genes essential for spermatogenesis are two genetic factors that contribute to NOA [8]. According to World Health Organization (WHO) guidelines, the term “azoospermia” is used when there are no sperm in the ejaculate, and “oligozoospermia” is used when the sperm concentration is <15 million sperm/mL of ejaculate (i.e., below the lower reference limit). There are three levels of oligozoospermia: mild (10 to 15 million sperm/mL), moderate (5 to 10 million sperm/mL), and severe (<5 million sperm/mL) [9,10].
The transition nuclear protein 2 (TNP2) and protamine 1 (PRM1) genes are both found on chromosome 16 (16p13.3), where they form a multigenic cluster with the protamine 2 (PRM2) gene [11]. TNP1 and TNP2, which have only one copy each, encode TP1 and TP2 proteins [12]. The two protamine genes and TNP2 have a close connection, suggesting they were formed through gene duplication and may still exhibit shared activities. During the stages of spermiogenesis, 90% of the chromatin-basic proteins are made up of TNPs [13]. At later stages of spermiogenesis, TNPs are shown to be essential for the replacement by protamine and sperm DNA condensation [14].
Synaptonemal complex protein 3 (SYCP3), on the 12q23.2 chromosome, is a crucial indicator of meiotic germ cell division throughout spermatogenesis. Some unexplained instances of azoospermia are thought to be caused by abnormalities in human synaptonemal complex development; thus, SYCP3 may be a tool for predicting the course of human spermatogenesis, particularly in infertile men [15,16]. Furthermore, it is thought that SYCP3 is mainly expressed in germ cells [15], and it is a testis-specific gene that functions as the central component of the lateral elements of the synaptonemal complex, which controls synapsis, sister chromatid cohesion, DNA binding to the chromatid axis, and recombination [17].
Moreover, the long arm of the Y chromosome contains the azoospermia factor (AZF) region. It is broken down into three subregions: AZFa, AZFb, and AZFc, and plays a crucial part in the genetics of male infertility [18]. Genes important to spermatogenesis and testicular development can be found in these locations. Microdeletions in this region cause male infertility and spermatogenetic abnormalities. The human Y chromosome is essential for the development and function of male germ cells as well as the determination of the human sex [19]. The proximal long arm of the Y chromosome (Yq) is where the AZFa subregion is found [20]. The primary gene of the AZFa area is DEAD-box helicase 3 and Y-linked (DDX3Y), which has been linked to the advancement of premeiotic germ cells and is expressed in the testis. This suggests that suppressing DDX3Y expression contributes to the loss of prenatal germ cells and to infertility [21,22]. The sY84 and sY86 markers are the first choice for sequence-tagged site (STS) primers in the AZFa region, their deletions suggest a very high probability of complete deletion of AZF, and NOA is frequently caused by complete deletion of the AZFa region [23]. Our study evaluated TNP2 and SYCP3 gene polymorphisms, AZFa microdeletions, and their gene expression levels in 100 patients with azoospermia.
Methods
1. Study participants
Patients with azoospermia, aged 28 to 40 years, were chosen for the examination of promoter region polymorphism in the TNP2 and SYCP3 genes and microdeletions in AZFa. All patients received a thorough andrological examination, which included a medical history and physical examination, semen analysis, scrotal ultrasonography, and hormone analysis. In addition, every patient was examined to determine whether their karyotype was normal (46XY). A comprehensive medical history and physical examination were conducted for the controls, and all were fertile and had normal children.
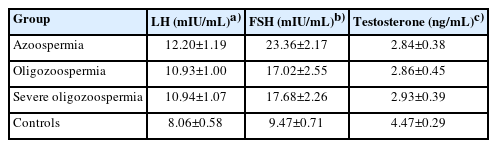
In total, 100 men were selected for gene expression analysis as patient group after undergoing bilateral microdissection testicular sperm extraction (mTESE) to obtain spermatozoa for intracytoplasmic sperm injection. Karyotyping and Y chromosomal microdeletion analyses and the blood levels of follicle-stimulating hormone (FSH), luteinizing hormone (LH), and testosterone were assessed before surgery. The patients were not on hormone treatment and had primary infertility. Mean±standard deviation values for the serum LH, FSH, and testosterone levels of participants are illustrated in Table 1. None of the 100 male controls had a history of mTESE or cryptorchidism. The study excluded patients with cystic fibrosis, chromosomal abnormalities, and Y chromosome microdeletion. The control group consisted of 100 men with normal spermatogenesis (>15.0×106 sperm/mL) according to the WHO criteria and confirmed fertility.
2. Sampling and DNA extraction
The research was approved by the local ethics committee (NO. 52/422370/1) and all subjects provided written informed consent. Blood samples from patients with azoospermia were collected to analyze gene polymorphisms and mutations. The salting-out DNA extraction technique was used to extract DNA samples from total blood samples.
To evaluate gene expression levels, frozen testis tissue was homogenized. Total RNA was extracted using the RNeasy Plus Universal Mini Kit (Qiagen) according to the manufacturer's protocol and stored at −80 °C. In-solution DNase digestion was used to eliminate DNA contamination. The concentration and purity of RNA were quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific) and confirmed by agarose gel electrophoresis. Template cDNA was synthesized from 1 μg of total extracted RNA using the Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific) simultaneously with oligo-dT and random hexamer primers for each reaction in an Eppendorf Mastercycler Gradient (Eppendorf AG).
3. Restriction fragment length polymorphism genotyping
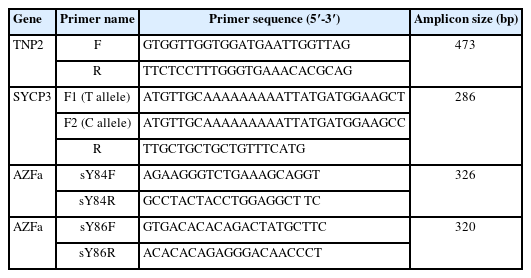
Polymerase chain reaction (PCR) amplification was performed using primer pairs (Table 2). PCR fragments were amplified as a 473bp fragment from the TNP2 gene, and the PCR conditions for TNP2 were as follows: 32 cycles of denaturation at 95 °C for 30 seconds, annealing at 57 °C for 1 minute, and extension at 72 °C for 30 seconds. The restriction fragment length polymorphism method was used for genotyping the TNP2 rs2857758 allele. The EarI restriction endonuclease enzyme was used to detect the CTCTTC sequence. The digestion reaction was carried out for 2 hours at 37 °C, and it digested 473-bp PCR products with altered G alleles into 353-bp and 120-bp fragments. The DNA fragments were isolated using 1.5% agarose gel electrophoresis and visualized in a gel documentation system by safe staining.
4. Allele-specific PCR
The presence of the T657C mutation in the SYCP3 gene was evaluated with the allele-specific (AS) PCR (ASPCR) technique. Two forward primers with variations in their 3' nucleotides were designed so that each was specific for one of the two variants (Table 2). They were combined with a common reverse primer into two PCR reactions. A 25-μL reaction mixture comprising 100 ng of DNA; 40 pmol of primers (SYCP3 F1, F2, and SYCP3 R); and 12.5 μL of 2x Taq PCR Master Mix was utilized for amplification. An initial melting phase of 94 °C for 5 minutes was followed by 35 cycles of 94 °C for 30 seconds, an annealing temperature starting at 60 °C for 30 seconds and 72 °C for 30 minutes for extension, and a final elongation step of 5 minutes at 72 °C. A gel documentation system was used to visualize the amplified DNA on 1.5% agarose gel with safe staining. The C and T alleles produced 286-bp bands due to the AS primers' reactions.
5. Multiplex PCR
Multiplex PCR was employed according to the standard protocol for analysis of the AZF region of the Y chromosome. The AZFa subregion was analyzed using STS primers (Table 1). The European Academy of Andrology recommends these STS primers, which can identify 90% of the deletions in AZF loci (sY84, sY86 for the AZFa region). A total volume of 25L was used for the PCR amplification, including 100 to 200 ng of human genomic DNA as a template, 2.5 mM deoxynucleotide triphosphates (2.5 mM each of 2'-deoxythymidine-5'-triphosphate [dTTP], 2'-deoxycytosine-5'-triphosphate [dCTP], 2'-deoxyguanosine-5'-triphosphate [dGTP], 2'-deoxyadenosine-5'-triphosphate [dATP]), oligonucleotide primers (0.1 to 2.0 μmol/L each of the forward and reverse primers), 10x Taq DNA polymerase assay buffer (Tris with 15 mM MgCl2), and 3 U of Taq DNA polymerase.
The conditions for thermocycling were standardized for the AZFa subregions, utilizing a TC-512 gradient thermocycler. PCR was performed on the samples in 35 cycles at 94 °C for 30 seconds, 53 °C for 45 seconds, and 72 °C for 60 seconds. Initial denaturation was performed at 94 °C for 5 minutes, followed by a final extension at 72 °C for 10 minutes. The PCR amplified products were stained with safe staining and identified on 2% agarose gel electrophoresis. Before loading the samples into the wells of the agarose gel, samples were mixed with gel loading dye in a ratio of 1:1.
6. Quantitative real-time PCR
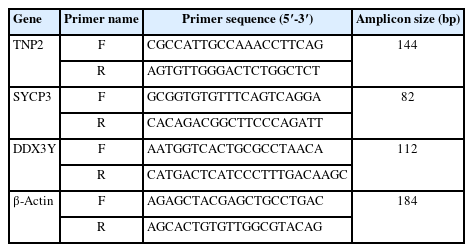
Expression analysis was carried out by quantitative real-time PCR (qRT-PCR) on cDNA libraries using primers specific for TNP2, SYCP3, DDX37 (located within the AZFa region), and the housekeeping gene β-actin (Table 3). The expression of β-actin was examined as an internal control for RNA isolation and RT-PCR efficiency. Initial denaturation for RT-PCR started at 95 °C for 8 minutes, followed by 40 cycles of denaturation at 95 °C for 10 seconds, annealing at 60 °C for 30 seconds, and extension at 72 °C for 30 seconds. To avoid primer dimer amplification, the melting curve was created by raising the temperature from 72 to 95 °C. The qPCR was performed in triplicate on 48-well plates in the StepOne-Plus RT-PCR System (Thermo Fisher Scientific/Applied Biosystems) using 1.0 μL of produced cDNA, 10 μL of the SYBR Green qPCR Master Mix (Applied Biosystems ABI/PE), and 7.0 μL of DNase/RNase-free water. One microliter of designed primers was used for the gene expression profile. The average cycles to threshold (CT) value was used for further analysis, and all RT-PCR cycles included non-template (cDNA) controls to rule out contamination. The comparative CT method was used to analyze relative gene expression (2-ΔΔCT). The 2-ΔΔCT parameter displays the expression level (expressed as a fold change) with respect to the housekeeping gene.
7. Statistical analysis
The chi-square test was used to compare allele and genotype frequencies between case and control groups. Statistical tests of significance and chi-square analysis were performed using R programming version 4.0.5 (R Foundation for Statistical Computing). Differences in the allelic and genotypic frequencies of rs2857758 for TNP2 and rs769825641 for SYCP3 between case and control groups were evaluated using the chi-square test with an odds ratio to express the risk of single-nucleotide polymorphisms (SNPs) for the disease of interest. The Fisher exact test was used for statistical testing when the allelic count was less than 5.
The gene expression results were verified as the mean±standard error of mean. The Mann-Whitney U test was used for analysis of non-normally distributed data. Statistical analysis was implemented using Prism 6 (GraphPad Software) and p-values <0.05 were deemed statistically significant. Furthermore, receiver operating characteristic (ROC) curve analysis and the area under the curve (AUC) were employed to assess the potential of these genes as diagnostic biomarkers.
Results
1. Gene polymorphisms
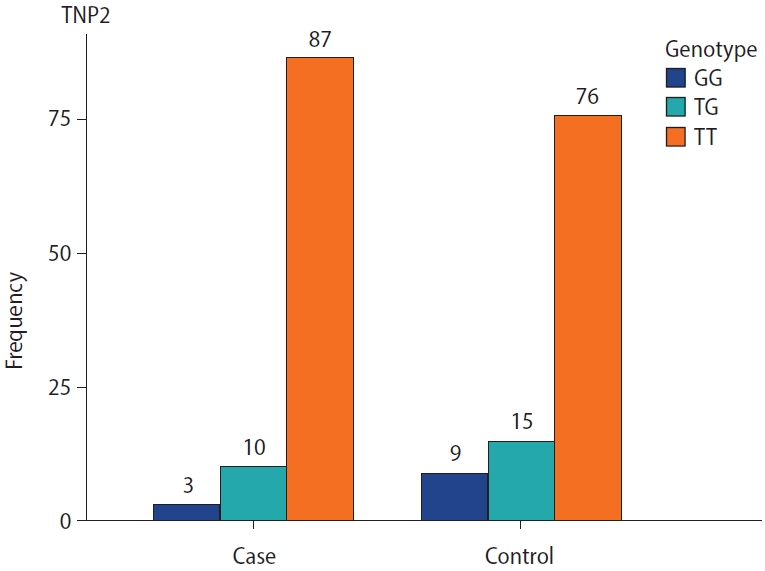
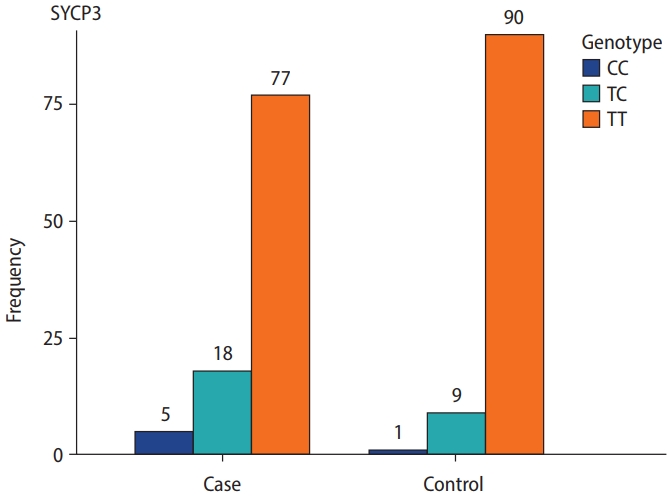
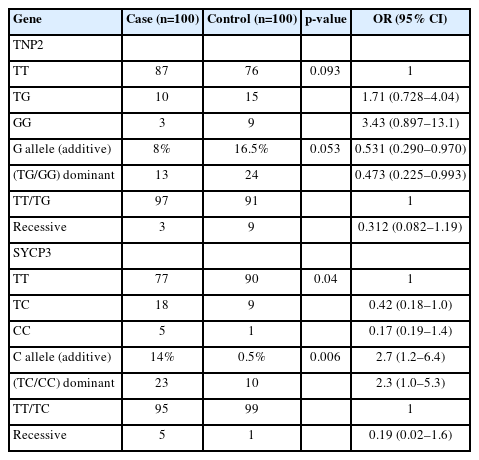
The genotype frequencies for the rs2857758 SNP of the TNP2 gene on chromosome 16 (16p13.13) were 87% for T/T, 10% for T/G, and 3% for G/G in sterile males, whereas the corresponding frequencies among the control group were 76%, 15%, and 9%, respectively (Figure 1). The altered allele frequency was higher among normal controls than in the case group. However, no significant difference (p=0.053) was found between infertile and fertile males (Table 3).
SYCP3 rs769825641 polymorphism, located on 12q23.2, was genotyped using ASPCR. Two PCR reactions were performed, one with forward primer 1 for the T allele and the other with forward primer 2 for the C allele, run side by side on an agarose gel. A 286-bp band indicated the presence of the specific allele. We observed that 167 (83.5%) cases and controls were homozygous TT, 27 (13.5%) were heterozygous TC, and six (3%) were homozygous CC. The genotyping frequency difference between the case and control groups was statistically significant (p=0.04). Additionally, the frequency of mutant alleles was higher in the sterile men than in the fertile male volunteers (Table 4, Figure 2).
Genotype and allelic frequency of TNP2 rs2857758 and SYCP3 rs769825641 in patients with azoospermia and a control group
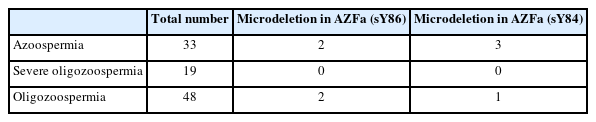
To validate the deletions in the AZFa region, the PCR reactions were repeated three times. The number of samples with deletions and their proportions are presented in Table 5. Deletions in the AZFa region were not found in patients with severe oligozoospermia. Among the 100 samples tested for the sY84 marker, four exhibited deletions in the AZFa region, and four showed deletions for the sY86 marker.
2. Expression analysis
The cDNA libraries from testis tissue were screened by PCR for the TNP2, SYCP3, and DDX3Y genes with their specific primers. Based on the Mann-Whitney U test, SYCP3 and DDX3Y showed altered expression in the testis tissue of infertile patients compared to fertile male controls. In contrast, no significant alteration was observed for TNP2 expression among the infertile males as compared to controls (Figure 3).
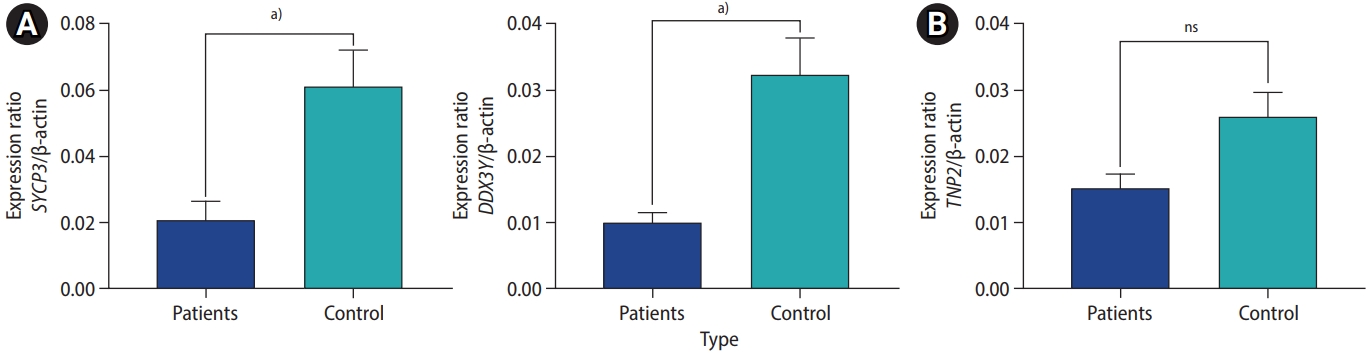
Gene expression levels in infertile patients compared to healthy subjects (controls). (A) synaptonemal complex protein 3 (SYCP3) and DEAD-box helicase 3 and Y-linked (DDX3Y) expression levels were significantly lower in patients than in controls (p<0.0001). (B) Transition nuclear protein 2 (TNP2) showed no significant alteration of expression in patients (p=0.095). ns, not significant. a)p<0.0001
3. Biomarker potential
ROC curve analyses were carried out to assess the diagnostic value of the TNP2, SYCP3, and DDX3Y genes for azoospermia. The results showed that SYCP3 and DDX3Y were potential biomarkers for azoospermia in fertile males, with AUCs of 0.772 and 0.720, respectively. However, TNP2 (AUC=0.568) showed no biomarker potential for diagnosing azoospermia (Figure 4).

Receiver operating characteristic curve analysis of: (A) synaptonemal complex protein 3 (SYCP3) with an area under the curve (AUC) of 0.722 (95% confidence interval [CI], 0.651 to 0.793; sensitivity=70%, specificity=61%), (B) DEAD-box helicase 3 and Y-linked (DDX3Y) with an AUC of 0.720 (95% CI, 0.649 to 0.790; sensitivity=67%, specificity=70%), and (C) transition nuclear protein 2 (TNP2) with an AUC of 0.568 (95% CI, 0.488 to 0.647; sensitivity=58%, specificity=56%).
4. Hormonal analysis
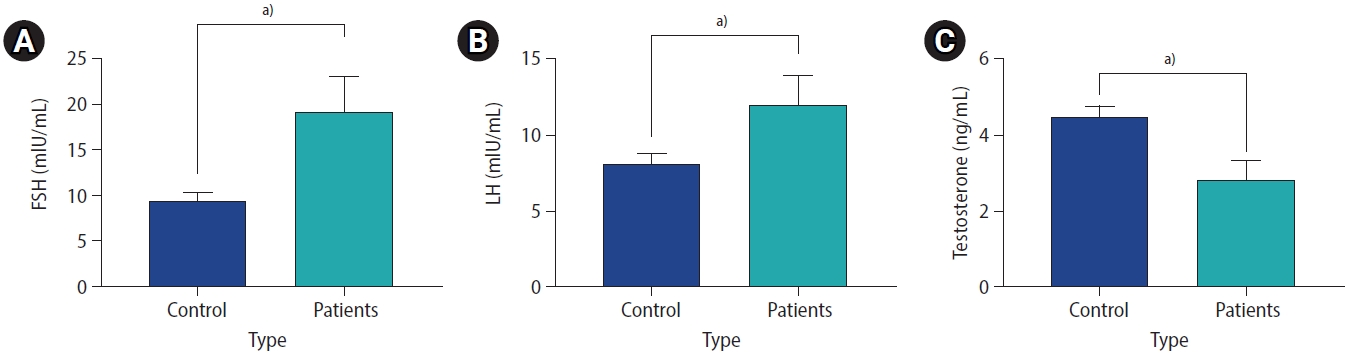
The analysis of hormone levels indicated that both LH and FSH levels were significantly lower in infertile patients than in healthy controls (p<0.001). In addition, testosterone levels were significantly lower (p<0.001) (Figure 5).
Discussion
Azoospermia, which affects roughly 1% of males, contributes significantly to male infertility [24]. Over the past 40 years, the diagnostic tools for identifying azoospermia have included standard tests for Y chromosome microdeletions, karyotype analysis, and a few monogenic investigations. Since the widespread adoption of whole-exome sequencing, increasingly many novel candidate genes associated with azoospermia have been identified, particularly in congenital HH [25].
The TNP gene is essential for maintaining the integrity of sperm DNA [8]. In chromatin remodeling and DNA compaction, TNP and PRM work together. In the postmeiotic phase of spermatogenesis, chromatin remodeling occurs as nucleosomal histones are initially replaced by TNPs (TNP1 and TNP2) and protamines (PRM1 and PRM2) to facilitate extensive chromatin condensation. Any disruption in the transition of either TNP1 or TNP2 may result in elevated oxidative stress, DNA strand breakage, and ultimately azoospermia due to activation of the apoptotic pathway [26]. In addition, it was discovered that mir-122a can reduce TNP2 expression during spermatogenesis. Arefnia et al. [27] demonstrated that the purpose of this miRNA is to inhibit gene expression by degrading the transcription of this gene.
One study concluded that the mRNA expression profile of TNP2 and the hormonal parameters in men with azoospermia were significantly associated with the successful retrieval of spermatozoa. Amjad et al. [26] concluded that the probability of spermatozoa retrieval in patients with azoospermia is elevated when the mRNA expression profile of TNP2 is increased. Heidari et al. [28] discovered that males with the CC genotype of the g.IVS1-26G>C SNP in the TNP2 gene have a greater chance of developing varicocele than men without this genotype.
The relative expression level of TNP2 in a study by Amor et al. [11] demonstrated a strong positive correlation with sperm count, progressive motility, and normal sperm morphology. They also suggested that smoking modifies the mRNA expression level of TNP2, which in turn alters normal sperm function [11].
In a study of genes associated with the VASA protein and protein-protein interactions, TNP2 and SYCP3 were identified as important genes causing infertility. Amirian et al. [17] observed that VASA and its interacting hub proteins may provide insights into the pathogenesis of aberrant germ cells and infertility. According to their findings, the TNP2 gene expression level was reduced in infertile cells compared to normal cells, while the SYCP3 gene expression level was increased [17].
Expression of the meiosis marker SYCP3 has been observed in spermatocytes [29]. According to Dhulqarnain et al. [15], osteocalcin (OCN) increased the expression of SYCP3 and enhanced spermatogenesis. Undercarboxylated OCN functions as a hormone, promoting insulin production, β-cell proliferation, and fertility [15]. Rybina et al. [30] suggested that one of the genetic causes of recurrent miscarriage is T657C polymorphism in the SYCP3 mutation of the synaptonemal complex.
A prevalent genetic cause of male infertility is Y chromosome microdeletion in the three subregions of AZF. The occurrence of co-microdeletions is also possible in these subregions. Y chromosomal microdeletions have been identified in a varying number of infertile males around the world [31]. If AZF microdeletions are found in a patient’s primary screening, further investigation is advised in the AZFa subregion [19,32]. Two genes, including ubiquitin specific peptidase 9 Y-linked (USP9Y) and DBY, also known as DDX3Y, are present in AZFa. Even in the earliest stages of development, it was significant to note that no DDX3Y expression was seen outside the tubules, indicating that the expression was only found in germ cells [33].
Y chromosomal microdeletions were found in a high percentage of infertile patients. Therefore, thorough evaluation of infertile males by semen analysis, hormonal assessment, and when necessary, karyotype analysis and physical examination may identify patients for whom a Y chromosome microdeletion study is required and also help to keep costs down [34]. Further studies are needed to confirm our results, and additional investigations are required to detect the effects of other polymorphisms and AZF region microdeletions on azoospermia.
This study investigated the genotype and allele frequencies of some gene polymorphisms (TNP2 rs2857758 and SYCP3 rs769825641) and microdeletions in the AZFa region to detect their associations with azoospermia in sterile males. The best control samples are individuals with confirmed fertility. However, in this study the control subjects had never been referred to infertility facilities for mTESE surgery; thus, we chose individuals with normal spermatogenesis samples as the control group.
Although we found no significant association between TNP2 rs2857758 polymorphism and azoospermia, the homozygous genotype TT for SYCP3 rs769825641 polymorphism was significantly associated with the incidence of azoospermia. Furthermore, analysis of Y chromosomal microdeletions can help clinicians counsel infertile patients before planning assisted reproductive techniques. The deletion of the AZFa region results in Sertoli cells, and there was an 8% deletion in the AZFa region using sY84 and sY86 markers.
In the current investigation, the gene expression profile was also examined. Significant downregulation of SYCP3 and DDX3Y expression was identified in the testis tissue of infertile males. In contrast, TNP2 downregulation was not statistically significant in patients with azoospermia compared to the control group. Moreover, ROC curve results suggested that SYCP3 and DDX3Y could be effective biomarkers for the diagnosis of azoospermia.
Notes
Conflict of interest
No potential conflict of interest relevant to this article was reported.
Author contributions
Conceptualization: MIIJ, ND, PB, RS. Data curation: MIIJ, ND, PB, RS. Formal analysis: MIIJ, ND, PB, RS. Methodology: MIIJ, ND, PB, RS. Visualization: MIIJ, ND, PB, RS. Writing-original draft: MIIJ, ND, PB, RS. Writing-review & editing: MIIJ, ND, PB, RS.