Nuclear localization of Obox4 is dependent on its homeobox domain
Article information
Abstract
Objective
Oocyte-specific homeobox 4 (Obox4) is preferentially expressed in oocytes and plays an important role in the completion of meiosis of oocytes. However, the Obox4 expression pattern has not been reported yet. In this study, we investigated the subcellular localization of Obox4 using a green fluorescent protein (GFP) fusion expression system.
Methods
Three regions of Obox4 were divided and fused to the GFP expression vector. The partly deleted homeodomain (HD) regions of Obox4 were also fused to the GFP expression vector. The recombinant vectors were transfected into HEK-293T cells plated onto coated glass coverslips. The transfected cells were stained with 4',6-diamidino-2-phenylindol and photographed using a fluorescence microscope.
Results
Mutants containing the HD region as well as full-length Obox4 were clearly localized to the nucleus. In contrast, the other mutants of either the N-terminal or C-terminal region without HD had impaired nuclear localization. We also found that the N-terminal and C-terminal of the Obox HD contributed to nuclear localization and the entire HD was necessary for nuclear localization of Obox4.
Conclusion
Based on the results of the present study, we demonstrated that the intact HD region of Obox4 is responsible for the nuclear localization of Obox4 protein in cells.
Introduction
The oocyte-specific homeobox (Obox) gene family consists of 6 members and is exclusively expressed in the ovaries, testes, and oocytes [1-3]. The Obox family was first identified as preferentially expressed genes in mammalian oocytes by in silico subtraction to identify oocyte-specific expressed sequence tags [1]. Obox genes contain a homeobox domain that encodes a peptide sequence with limited homology to other homeodomain protein sequences. Homeobox genes play important roles in many developmental processes by regulating gene transcription [4,5]. Mice lacking the Obox6 gene grow without embryonic development abnormalities and with normal fertility, suggesting functional redundancy among the Obox family members [2].
It has been found that the expression of all Obox family members except Obox4 was ovary-specific but the expression of Obox4 was preferentially transcribed in the testis [1]. However, we previously found that Obox4 was expressed abundantly in the oocytes, ovaries, and testes [6]. During oocyte maturation, prophase I arrested germinal vesicle (GV) stage oocytes undergo germinal vesicle breakdown and progress through meiosis to metaphase II (MII), and are arrested again at MII [7]. By RNA interference and overexpression analysis, we found that Obox4-deficient oocytes induced abnormal metaphase I arrest during in vitro maturation and the arrest was resumed by overexpression of Obox4 in the treatment of 3-isobutyl-I-methyl-xanthine, a phosphodiesterase inhibitor, which maintains an intact GV state [6]. These results indicate that Obox4 plays an important role in the cyclic adenosine monophosphate (cAMP)-dependent signaling pathways that maintain GV arrest. In other cells, it was reported that Obox4 is expressed in mouse embryonic stem cells and induces the differentiation of the cells through the regulation of the histone gene family [8].
The Obox4 protein can be divided into three regions: the N-terminus, the homeodomain (HD) region containing the putative DNA binding motif, and the C-terminus. However, in spite of the biological importance of Obox4 in terms of oocyte maturation, little is known about of the expression pattern of Obox4 and the related role of the functional domains. Therefore, in this study, we investigated the cellular distribution of Obox4 using a variety of fusion proteins with green fluorescent protein (GFP) constructed by deletion mutagenesis. We found that the intact HD region of Obox4 is required for its complete nuclear localization.
Methods
1. Plasmids constructs
The full-length cDNA of Obox4 obtained from the cDNA of mouse oocytes was amplified with primers (5'-GCGCAGATCTCGATGTCCAAAGATTCCTCCTTG-3'and 5'-GCGCTCGAGCTATAGAGACATCATGGCATC-3') and ligated into BglII and SalI sites of pEGFP-C2 (Clonech, Palo Alto, CA, USA). The N-terminus, N-terminus plus HD, C-terminus, and C-terminus plus HD regions fused with GFP were generated by ligating each domain into the same sites of the enhanced green fluorescent protein (pEGFP) vector. The GFP fusion proteins with partially deleted HD sequences were also generated by the same method as above using the specific primers. All recombinant vectors were confirmed by sequencing.
2. Cell culture and transfection
HEK-293T cells were cultured with Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco Invitrogen, Carlsbad, CA, USA), 100 U/mL of penicillin, and 100 g/mL of streptomycin. The cells were plated onto glass coverslips coated by 0.1 mg/mL poly-d-lysine in 4-well plates and grown overnight. The cells were transfected with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Briefly, a total of 1×105 cells were transfected using 1 g of each recombinant vector and 2.5 L of Lipofectamine 2000 per well. After 5 hours of incubation, 0.5 mL of growth medium was added to the cells. The next day, the medium was changed. After 24 hours of incubation, the cells were analyzed. Following fixation in 4% paraformaldehyde and staining of nuclear DNA with 4',6-diamidino-2-phenylindol (DAPI), the coverslips were mounted on glass slides. The cells were photographed using fluorescence microscopy (Eclipse TE-2000 U, Nikon, Tokyo, Japan).
Results
To examine the cellular localization of Obox4 and to delineate the functional nuclear localization regions, we analyzed the domain sequence of Obox4 using the online Motif search program (http://www.genome.jp/tools/motif). The region between amino acids 92 and 152 was predicted to be a HD region having a similarity score 926 when compared to the consensus HD sequence (Figure 1A). To confirm the prediction, we first generated a recombinant vector expressing full-length Obox4 fused to the C-terminus of enhanced green fluorescent protein (EGFP) in the pEGFP vector (EGFP-Obox4-FL). In transiently transfected HEK-293T cells, full-length Obox4 was strictly localized to the nucleus (Figure 1B).

Nuclear localization of full-length Obox4 fused to GFP. (A) Schematic representation of Obox4 protein structure. The HD sequence of Obox4 was aligned with the consensus sequence of HD. (B) Schematic representation showed GFP vector control and GFP-fused full-length recombinant Obox4. EGFP vector and full-length Obox4 fused to GFP were transfected into HEK-293T cells. The transfected cells were counter-stained with DAPI for nuclear staining and photographed by fluorescence microscopy. Bar=25 µm. Obox, oocyte-specific homeobox; GFP, green fluorescent protein; HD, homeodomain; EGFP, enhanced green fluorescent protein; DAPI, 4',6-diamidino-2-phenylindol.
In order to examine the domain responsible for nuclear localization, we divided Obox4 into three regions, and then we generated recombinant vectors expressing each domain of Obox4: the N-terminal region (Figure 2A: EGFP-Obox4-N, amino acids 1-91), the HD (Figure 2B: EGFP-Obox4-HD, amino acids 92-152), and the C-terminal region (Figure 2C: EGFP-Obox4-C, amino acids 153-453). The recombinant vectors containing N-terminal or C-terminal regions without HD were localized to the cytoplasm as well as the nucleus (Figure 2A, C), while the recombinant vectors containing HD alone were exclusively localized to the nucleus (Figure 2B). These results imply that the region responsible for nuclear localization resides in the HD. To confirm this result, we generated recombinant vectors expressing the HD plus the N-terminal (Figure 2D: EGFP-Obox4-NH, amino acids 1-152) or C-terminal regions of Obox4 (Figure 2E: EGFP-Obox4-CH, amino acids 92-453). We found that both of these recombinant proteins were clearly localized to the nucleus (Figure 2D, E). This result indicates that nuclear localization of Obox4 only requires the functionally responsible sequence within the HD.
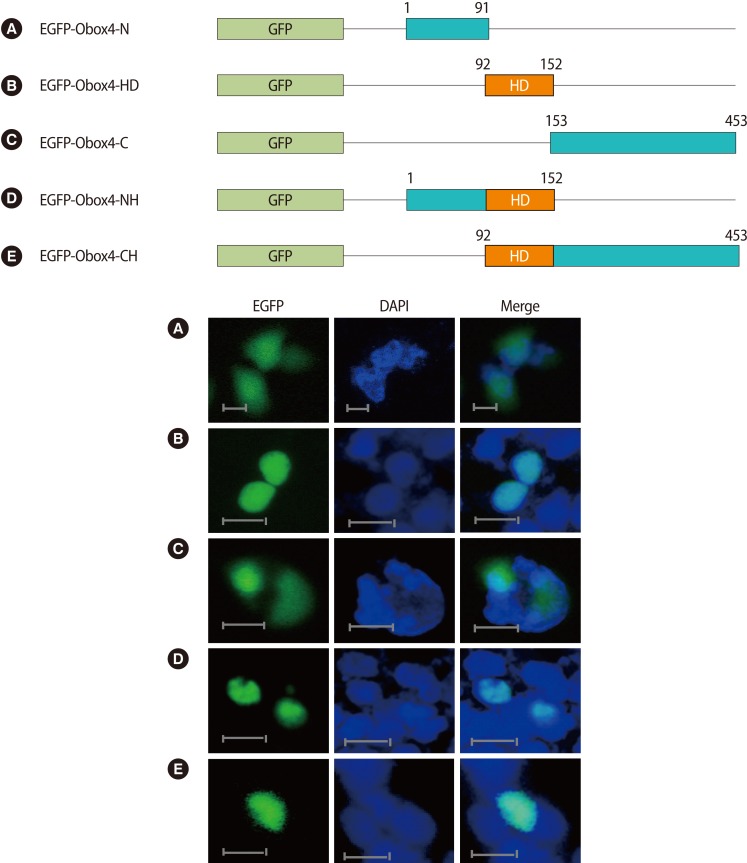
Cellular localization of GFP-fused truncated Obox4 containing the N-terminal, the C-terminal, or the HD alone and their combinations. Schematic representation showed that GFP-fused each domain of the Obox4 proteins. Each recombinant vector was transfected into HEK-293T cells. The transfected cells were counterstained with 4',6-diamidino-2-phenylindol for nuclear staining and photographed by fluorescence microscopy. (A) EGFP-Obox4-N, amino acids 1-91. (B) EGFP-Obox4-HD, amino acids 92-152. (C) EGFP-Obox4-C, amino acids 153-453. (D) EGFP-Obox4-NH, amino acids 1-152. (E) EGFP-Obox4-CH, amino acids 92-453. Bar=25 µm. EGFP, enhanced green fluorescent protein; GFP, green fluorescent protein; HD, homeodomain; N, N-terminal; C, C-terminal; NH, N-terminal plus homeodomain; CH, C-terminal plus homeodomain.
To further investigate the elements necessary and sufficient for nuclear localization in the HD, we performed sequence analysis using the PSORTII prediction program (http://psort.hgc.jp). Though we could not find the nuclear localization signal motif in Obox4, the program predicted the nuclear localization of Obox4 with high reliability. Thus, based on previous reports that the nuclear localization signals are located in both ends of the HD of hNANOG and hNK2.2 [9,10], we generated several recombinant vectors expressing the intact HD (Figure 3A: EGFP-Obox4-HD, amino acids 92-152) and the HD without the N-terminal (amino acids 92-97) or C-terminal (amino acids 145-152) part and without both parts (Figures 3B: EGFP-Obox4-98HD, amino acids 98-152; Figures 3C: EGFP-Obox4-144HD, amino acids 92-144; and Figures 3D: EGFP-Obox4-98/144HD, amino acids 98-144). Expression deficient in either the N-terminal or C-terminal part of the HD resulted in less efficient nuclear localization compared to that of the intact HD (Figure 3B, C). Deletion of both ends of the HD also showed dispersed localization of the recombinant protein (Figure 3D). Furthermore, we divided the HD into two half parts and generated recombinant vectors expressing each half part (Figure 3E: EGFP-Obox4-FirstHD, amino acids 92-122; and Figure 3F: EGFP-Obox4-SecondHD, amino acids 123-152). Expression of each half part of the HD resulted in less strict nuclear localization of the proteins compared to the intact HD (Figure 3E, F). These results suggested that the whole HD region including both ends of the HD were required for efficient and complete nuclear localization of Obox4.
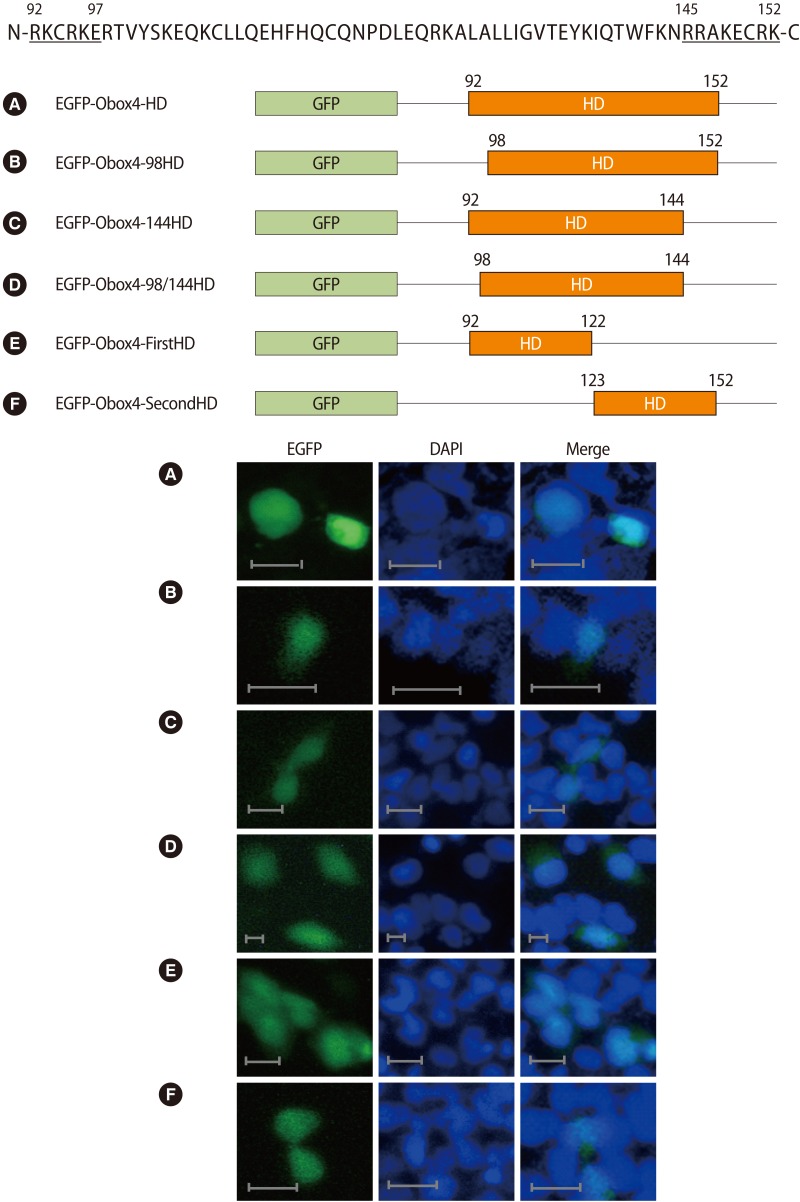
Deletion analysis of the Obox4 HD. The HD sequence of Obox4 is shown at the top of the figures with capitalized letters and the deleted N-terminal and C-terminal end parts of the HD are underlined. A schematic representation of the partly deleted mutants of Obox4 HD fused to GFP is shown (A-F). Each recombinant vector was transfected into HEK-293T cells. The transfected cells were counterstained with DAPI for nuclear staining and photographed by fluorescence microscopy. Bar=25 µm. EGFP, enhanced green fluorescent protein; GFP, green fluorescent protein; HD, homeodomain; N, N-terminal; C, C-terminal; N, N-terminal; C, C-terminal; DAPI, 4',6-diamidino-2-phenylindol.
Discussion
The homeobox domain was first identified in a number of drosophila homeotic and segmentation proteins, but is now known to be found in many other animals, including vertebrates [11]. Generally, the HD is a highly conserved domain that encodes a protein domain having a capability to bind DNA, and homeobox genes encode transcription factors that activate or repress the transcription of downstream target genes.
The full-length and the HD of Obox4 were clearly localized to the nucleus and any regions without the HD were not strictly localized to the nucleus (Figures 1, 2). These results indicated that the motif for nuclear localization was exclusively present in the HD. Recombinant Obox4 without the HD proteins showed dispersion throughout the cytoplasm as well as nuclear localization. This could have been caused either by passive diffusion from the nucleus to the cytoplasm or by an inefficient nuclear import process. Nuclear proteins transported by the nuclear import machinery mediated by nuclear localization signals are composed of highly basic amino acid residues [12,13]. Although sequence alignment between the HD of Obox4 and that of hNANOG shows no significant homology in the motif of nuclear localization signals, the deletion of both ends of the Obox4 HD impaired nuclear localization (Figure 3). Furthermore, each half part of the HD was necessary for nuclear localization. These results suggested that there is a region responsible for nuclear localization of Obox4 throughout the entire HD, although a certain motif cannot yet be specified.
HD proteins are usually crucial transcription factors for cell differentiation, cell proliferation, and tissue development. Intriguingly, their HD signature structure is important for both their DNA binding ability and their nucleocytoplasmic trafficking [14]. The accurate nucleocytoplasmic distribution of these proteins is essential for their functioning [15]. hNANOG, for example, has a nuclear export signal in its HD and it controls the nucleocytoplasmic translocation of the protein [16]. A mutant of hNANOG containing only a nuclear export signal motif was located predominantly in the cytoplasm. The mutants of Obox4 containing truncated HD showed a cellular location dispersed from the nucleus to the cytoplasm (Figure 3), suggesting that the Obox4 HD would not have contained the nuclear export signal or that it had been broken by deletion analysis. HD has three alpha helices (helix I, II, and III) forming a helix-turn-helix motif and DNA recognition [17]. Depending on the characteristics of proteins, the crucial motif for nuclear localization resided either in helix I or helix III [18,19], and mouse NKX2.2 has two motifs in both ends of the HD, helix1 and helix III [10]. Although Obox4 shares no significant homology with hNANOG and NKX2.2, both ends of the HD contributed to the nuclear localization of Obox4. Further studies are needed to disclose the structure and function of the Obox4 HD.
In this study, we report for the first time that Obox4 was localized to the nucleus of HEK-293T cells and a requirement for the localization was the entire region of the HD of Obox4. We used a GFP fusion expression system to investigate the subcellular localization of Obox4. As we previously reported, Obox4 is specifically expressed in mouse oocytes and plays important roles in oocyte maturation [6]. Therefore, further studies are required to define the minimal and fully functional nuclear localization signal of Obox4 and other cellular localization signals such as nuclear export signals to understand the multiple roles of Obox4 in cells and eventually in oocytes in particular.
Notes
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (20100022600) and by the Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2009-0093821).
No potential conflict of interest relevant to this article was reported.