Vitrification, in vitro fertilization, and development of Atg7 deficient mouse oocytes
Article information
Abstract
Objective
Autophagy contributes to the clearance and recycling of macromolecules and organelles in response to stress. We previously reported that vitrified mouse oocytes show acute increases in autophagy during warming. Herein, we investigate the potential role of Atg7 in oocyte vitrification by using an oocyte-specific deletion model of the Atg7 gene, a crucial upstream gene in the autophagic pathway.
Methods
Oocyte-specific Atg7 deficient mice were generated by crossing Atg7 floxed mice and Zp3-Cre transgenic mice. The oocytes were vitrified-warmed and then subjected to in vitro fertilization and development. The rates of survival, fertilization, and development were assessed in the Atg7 deficient oocytes in comparison with the wildtype oocytes. Light chain 3 (LC3) immunofluorescence staining was performed to determine whether this method effectively evaluates the autophagy status of oocytes.
Results
The survival rate of vitrified-warmed Atg7f/f;Zp3-Cre (Atg7d/d) metaphase II (MII) oocytes was not significantly different from that of the wildtype (Atg7f/f) oocytes. Fertilization and development in the Atg7d/d oocytes were significantly lower than the Atg7f/f oocytes, comparable to the Atg5d/d oocytes previously described. Notably, the developmental rate improved slightly in vitrified-warmed Atg7d/d MII oocytes when compared to fresh Atg7d/d oocytes. LC3 immunofluorescence staining showed that this method can be reliably used to assess autophagic activation in oocytes.
Conclusion
We confirmed that the LC3-positive signal is nearly absent in Atg7d/d oocytes. While autophagy is induced during the warming process after vitrification of MII oocytes, the Atg7 gene is not essential for survival of vitrified-warmed oocytes. Thus, induction of autophagy during warming of vitrified MII oocytes seems to be a natural response to manage cold or other cellular stresses.
Introduction
Macroautophagy (referred to hereafter as autophagy) is an intracellular catabolic mechanism that degrades old or dysfunctional cellular components through lysosomes [1]. Catabolic products are then recycled to generate new membranes and macromolecules to maintain normal cell function [2]. This process is thought to occur at a basal level in all cells while certain signals upregulate or suppress this process. Many inducers of autophagy have been identified, including starvation, deprivation of growth factors or hormones, and temperature stress [234]. The process mostly relies on a well-characterized multistep pathway involving proteins encoded by various autophagy genes (Atg). Among these, Atg5 and Atg7 are widely recognized as upstream effectors whose actions are required for autophagosome formation. Atg5 forms a complex with Atg12, and this complex acts as an E2-like enzyme mediating the lipidation of Atg8 (light chain 3, LC3) protein. Lipidated LC3, LC3-II, is the phosphoethanolamine conjugated form and is incorporated into the autophagosomal membrane. Lipidation of LC3, i.e., the conversion of LC3-I to LC3-II, is often used as a marker of autophagic flux [1]. Atg7 is an E1-like enzyme that conjugates with LC3 and Atg12 during the biogenesis of autophagosomes. Therefore, it sits at the upstream of autophagic activation [56]. The mouse model of floxed Atg7 is now widely used to investigate the effects of defective autophagy in various tissues and organs [78]. Zona pellucida protein 3 (Zp3) gene encodes an oocyte-specific protein expressed during oogenesis [9]. The Zp3-Cre transgenic mouse model is used to drive deletion of the floxed gene in oocytes specifically [10].
We have previously showed that mouse oocytes show a heightened autophagic response during warming after vitrification [11]. Along with several ultrastructural and cellular changes [1213], autophagy is recognized as a cellular response to various stresses to oocytes and embryos [1114]. However, the role of autophagic activation during warming is unclear, as vitrified-warmed oocytes are generally considered normal with respect to rates of survival, fertilization, and subsequent development in mice [15]. Thus, autophagic activation observed during warming could be a cell response associated with restoration of potential cellular damage done by temperature stress. Alternatively, autophagy could be activated as a natural response to cold stress but may not have a necessary function for survival. Thus, it remains to be determined whether a complete absence of autophagy has any effect on the survival, fertilization, or development of vitrified-warmed oocytes. In this work, we aim to address two aspects of autophagy in mouse oocytes: first, the consequences of a complete absence of Atg7, a seminal gene in autophagy, in vitrified-warmed oocytes, and second, assessment of LC3 immunofluorescence staining as a reliable method of monitoring autophagy in oocytes.
Methods
1. Mice
All mice were maintained in strict accordance with the policies of the Konkuk University Institutional Animal Care and Use Committee (IACUC). This study was approved by the Konkuk University IACUC (approval no. KU14099). Atg7 floxed mice (Atg7f/f mice, C57BL/6) [7] were obtained from the RIKEN BioResource Center (Ibaraki, Japan). Zp3-Cre transgenic mice (C57BL/6) were purchased from Jackson Laboratory (Bar Harbor, ME, USA). To produce the Atg7f/f;Zp3-Cre mice, Atg7f/f female mice were mated with Atg7f/+;Zp3-Cre male mice, and the litters were selected as either experimental or control depending on the genotype (Table 1). To improve reproductive performance, the mice were crossed onto an ICR background. Deletion of the Atg7 gene in the oocytes of the Atg7f/f;Zp3-Cre mice was confirmed by reverse transcription polymerase chain reaction (RT-PCR) (Table 1).
2. Collection of matured oocytes
Atg7f/f female mice were intraperitoneally injected with 7.5 IU of pregnant mare's serum gonadotropin (Sigma-Aldrich, St. Louis, MO, USA) to induce folliculogenesis, followed by injection of 7.5 IU of human chorionic gonadotropin (Sigma-Aldrich) 48 hours later to induce superovulation. Cumulus-oocyte complexes were collected from the oviducts 13 to14 hours post-injection. The metaphase II (MII) oocytes were retrieved in Quinn's Advantage Medium with HEPES (Sage In Vitro Fertilization, Trumbull, CT, USA) containing 20% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA) at 37℃. The cumulus cells were then removed by using 300 µg/mL hyaluronidase (Sigma-Aldrich).
3. Vitrification and warming procedure
The vitrification procedure that was used is described in [11]. The oocytes were pre-equilibrated with a solution of 7.5% ethylene glycol (EG) (Sigma-Aldrich), 7.5% dimethyl sulfoxide (DMSO) (Sigma-Aldrich) and 20% FBS for 2.5 minutes. They were then equilibrated with 15% EG, 15% DMSO, 20% FBS, and 0.5 M sucrose (Fisher Scientific, Fair Lawn, NJ, USA) for 20 seconds. The equilibrated oocytes was loaded onto a Cryotop (Kitazato, Shizuoka, Japan) and immersed in liquid nitrogen (LN2). The vitrified oocytes were stored in LN2 for 2 weeks. After 2 weeks, they were warmed with sucrose solutions of decreasing concentrations (0.5, 0.25, 0.125, and 0 M) and 20% FBS for 2.5 minutes each. The vitrified-warmed oocytes were then cultured for 1 to 2 hours in medium with HEPES containing 20% FBS for recovery.
4. In vitro fertilization and embryo culture
In vitro fertilization (IVF) was performed according to the procedure described by Cha et al. [16]. Epididymal sperm waom 10–11 week-old male ICR mice. For capacitation, a drop of sperm suspension was added to Quinn's Advantage Fertilization Medium human tubal fluid (HTF) (Sage) containing 10% substitute protein serum (SPS, Sage), and incubated for 90 minutes at 37℃ in 5% CO2. For IVF, the capacitated sperm suspension was added to oocytes in HTF containing 10% SPS. After 6 hours, the oocytes were washed four times with HTF containing 10% SPS and finally with amino acid-supplemented potassium simplex optimized medium (KSOM-AA) medium. They were then cultured in KSOM-AA drops overnight. Fertilization was confirmed 24 hours later by observing the two-cell embryos. They were cultured for 5 days to assess embryonic development.
5. RNA extraction and RT-PCR
Total RNA from the MII oocytes (50 oocytes/sample) and ovaries were extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Rabbit α-globin RNA (10 pmol/sample, Sigma-Aldrich) was used as an external control [11]. To isolate minute amounts of RNA from oocytes, 3 M sodium acetate (pH 5.2) and glycogen were used in a modified protocol. The resuspended RNA was treated with RNase-free DNase I (Roche, Mannheim, Germany) for 20 minutes at room temperature to remove any residual genomic DNA. Total RNA was then reverse transcribed using Moloney murine leukemia virus reverse transcriptase (BEAMS Biotechnology, Seongnam, Korea) and random hexamer primers (Roche) for cDNA synthesis. The cDNA samples were used as templates for PCR analysis. PCR was performed with Prime Taq Premix (2×) (Genet Bio, Daejeon, Korea). Ribosomal protein L7 was used as an internal control. The RT-PCR was repeated four times with independent sample sets. The sequences of primers used for PCR analysis are given in Table 1.
6. Immunofluorescence staining and confocal microscopy
Oocytes were fixed and permeabilized with 4% paraformaldehyde with 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 20 minutes, and washed twice with 0.1% Triton X-100 in PBS for 5 minutes each. The oocytes were then blocked in a 2% bovine serum albumin (BSA)/PBS drop for 1 hour. They were then incubated with a primary antibody drop containing anti-LC3 (rabbit polyclonal, 1:400, Cell Signaling Technology, Danvers, MA, USA) in 2% BSA/PBS overnight at 4℃. The oocytes were washed three times in 2% BSA/PBS, and incubated with an Alexa Fluor 488 chicken anti-rabbit secondary antibody (1:250, Molecular Probes, Waltham, MA, USA) for 40 minutes. The oocytes were then counterstained with TO-PRO-3-iodide (1:250, Molecular Probes), washed in 2% BSA/PBS, directly placed on a glass slide, and covered with a glass coverslip that was sealed with transparent nail polish. Rabbit IgG was used as a mock control. Immunofluorescence images were obtained using an LSM710 confocal microscope (Carl Zeiss, Oberkochen, Germany), and analyzed using the Zen 2009 Light Edition (Carl Zeiss), a platform associated with the confocal microscope. For quantification of the LC3 puncta, each oocyte was imaged at four planes (3 µm intervals) using an LSM710 confocal microscope. The number of LC3-positive puncta was quantified by using the count tool in Image J ver. 1.50b (National Institutes of Health, Bethesda, ME, USA, http://imagej.nih.gov/ij/).
7. Statistical analyses
All data were graphed using GraphPad Prism ver. 5 (GraphPad, La Jolla, CA, USA). Statistical analyses were performed using the Student t-test (one-tailed). The criteria for statistical significance were p<0.05 and p<0.001.
Results
1. Confirmation of Atg7 deletion in oocytes from Atg7f/f;Zp3-Cre mice
To examine whether defective autophagy affects the survival, fertilization, and development of vitrified-warmed mouse oocytes, we used Atg7 floxed mice [7] bred with Zp3-Cre transgenic mice [10]. Atg7f/f female mice were mated with Atg7f/+;Zp3-Cre male mice, the litters of which were selected as experimental (Atg7f/f;Zp3-Cre, referred hereafter as Atg7d/d) or control (Atg7f/f) by genotyping (Figure 1A). Deletion of the Atg7 gene in Atg7d/d MII oocytes was confirmed by absence of Atg7 mRNA expression in MII oocytes in the RT-PCR analysis. As shown in Figure 1B, Atg7 mRNA expression was maintained in the entire ovary of Atg7d/d mice whereas their oocytes completely lost their expression. As expected, Cre mRNA expression was only noted in the MII oocytes of Atg7d/d mice. This suggests a complete deletion of Atg7 in the oocytes specifically.

Survival, fertilization, and development of vitrified-warmed oocytes from Atg7f/f; Zp3-Cre mice. (A) Oocyte-specific deletion of the Atg7 gene in Atg7f/f;Zp3-Cre mice (Atg7d/d) was confirmed. Primers used for genotyping are shown in Table 1. (B) RT-PCR results of Atg7 and Cre in oocytes from Atg7d/d and Atg7f/f mice. RNA from the ovaries (not reverse transcribed) was used as a negative control. rpl7 expression was examined as the loading control. Primers used for RT-PCR are shown in Table 1. (C) Survival rates of Atg7d/d and Atg7f/f oocytes after vitrification and warming. There was no significant difference in the survival rates of the Atg7 deficient oocytes (p=0.500). The values represent the mean±standard deviation (n=4). Statistical significance was assessed by the Student t-test. (D) Fertilization and developmental rates of vitrified-warmed Atg7d/d and Atg7f/f oocytes. The values represent the mean±standard deviation (n=3–4). The table shows the total number and percentage of oocytes that were used in the barograms. Statistical significance was assessed by the Student t-test. Zp3, zona pellucida protein 3; rpl7, ribosomal protein L7; RT-PCR, reverse transcription polymerase chain reaction; RT, reverse transcription; no-RT, no reverse transcription. *p<0.05; **p<0.001.
2. Survival of oocytes from Atg7d/d mice after vitrification
The survival rates of MII oocytes after vitrification and warming in Atg7d/d and Atg7f/f mice were evaluated (Figure 1C) and found to be similar (Atg7f/f:Atg7d/d=87.6%:86.1%, p=0.500). This indicates that the absence of autophagic activation in vitrified-warmed oocyte does not adversely affect oocyte survival after vitrification.
3. Fertilization and developmental rates of oocytes from Atg7d/d mice after vitrification-warming
The surviving Atg7f/f and Atg7d/d oocytes were inseminated, and their fertilization and developmental rates were assessed. As shown in Figure 1D, fresh oocytes from the Atg7d/d mice showed significantly lower rates of fertilization (Atg7f/f:Atg7d/d=63.1%:36.6%, p=0.047) and development (Atg7f/f:Atg7d/d=79.2%:50.0%, p<0.001) compared with the control oocytes. This is in concordance with a previous study where Atg5-deficient oocytes showed reduced rates of embryonic development [17]. In the vitrified-warmed groups, the fertilization rate was comparable between the two groups (Atg7f/f:Atg7d/d=42.5%:50.6%, p=0.289). Interestingly, vitrified-warmed Atg7d/d oocytes showed a higher developmental rate to the blastocyst stage than fresh Atg7d/d oocytes (fresh:vitrified-warmed=50.00%:77.6%, p<0.001).
4. Expression of LC3 in Atg7d/d oocytes
Among the many molecules involved in the autophagy pathway, LC3 is widely used to monitor dynamic autophagic flux by western blotting, immunofluorescence staining, or live imaging [1]. In our previous work showing activation of autophagy in oocytes during warming [11], we used GFP-LC3 transgenic oocytes, which enable observation of GFP-LC3 puncta formation by live imaging [18]. This method provides a consistent result when observing in vivo LC3 flux. Western blotting also provides a reliable readout of autophagic flux, as the conversion from the I-form to the II-form is easily distinguishable due to differential migrations [1]. On the other hand, observation by LC3 immunofluorescence staining requires caution [19]. While LC3-II proteins generally exhibit large puncta around autophagosomes, both LC3-I and LC3-II can be incorporated into protein aggregates in the cytoplasm [19], leading to misinterpretation under certain experimental conditions. Thus, we examined whether LC3 immunofluorescence staining can be reliably used as a monitoring method of autophagy in mouse oocytes using Atg7d/d oocytes. We examined LC3 localization with the anti-LC3 antibody by immunofluorescence staining. As shown in Figure 2A, large LC3 puncta were notable in Atg7f/f oocytes. Puncta were also noted in Atg7d/d oocytes, but the size and number were notably lower. For each oocyte, we counted the number of LC3-positive puncta and plotted them as shown in Figure 2B. The much lower number of puncta in the Atg7d/d oocytes suggests that LC3 immunostaining is a dependable monitoring method of autophagy in oocytes. Of note, a background level of LC3-positive signals is expected in Atg7 deficient tissues and cells and should therefore be interpreted with caution.
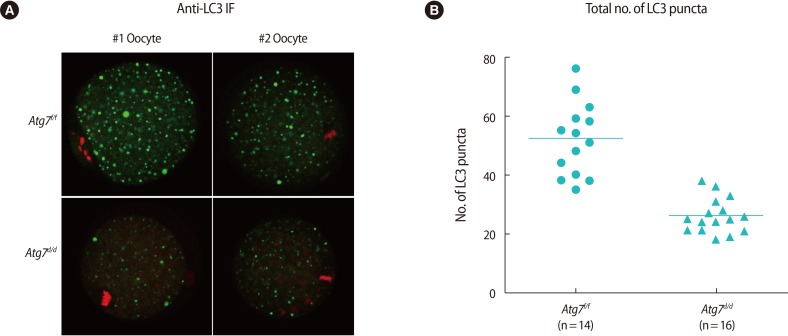
Immunofluorescence staining of LC3 in Atg7f/f MII oocyte. (A) Two representative oocytes are shown (#1 and #2). MII oocytes from Atg7f/f and Atg7d/d mice were vitrified-warmed and subjected to immunofluorescence staining with the anti-LC3 antibody. Alexa Fluor 488 (Molecular Probes, Waltham, MA, USA)-conjugated chicken anti-rabbit IgG was used as the secondary antibody (green), and nuclei were counter-stained with TO-PRO-3-iodide (Molecular Probes) (red). Images were obtained by confocal microscopy (shown at ×80). (B) For each oocyte, four images were captured at 3 µm intervals, and green dots were counted by Image J software (National Institutes of Health, Bethesda, ME, USA). The number of green dots read in four planes was added and plotted by using GraphPad Prism (ver. 5, GraphPad, La Jolla, CA, USA) software. The horizontal lines represent the mean numbers. LC3, light chain 3; MII, metaphase II.
Discussion
Autophagy, a subcellular self-eating process, occurs in normal physiology and various disease conditions. The autophagic on-rate can increase by diverse insults and stimuli to cells, and the list of autophagic inducers is ever expanding [2]. Unveiling new inducers sheds light on the underlying regulatory mechanisms. In mammalian oocytes and embryos, several inducers of autophagy have been identified, mostly the adverse effectors [14]. Vitrification involves rapid temperature change that would affect the configuration of macromolecules, and autophagy is induced in mouse oocytes during recovery after vitrification [11]. In this work, we further show that Atg7 is dispensable for cold stress exposed to oocytes during vitrification. Consistent with a study showing the role for autophagy in preimplantation embryonic development, as shown with Atg5 deficient oocytes [17], Atg7d/d oocytes also exhibit reduced developmental rates. However, reduced fertilization rates were not reported in the case of Atg5-deficient oocytes [17], suggesting that Atg7 may serve a more expanded role in the same pathway. With respect to vitrification, we observed that rates of survival, fertilization, and development were not further affected by it (Figure 1D). Thus, as we assumed in our previous work, autophagic induction during warming is likely a natural response to cold stress. Interestingly, vitrified-warmed Atg7d/d MII oocytes seemed to develop better to the blastocyst stage (Figure 1D). While this observation is interesting, it remains elusive what brought about such an outcome, which warrants further investigation. We recently showed that rapamycin treatment of MII oocytes increased autophagy and had an adverse effect on fertilization and development [20]. Taken together with our findings herein, suppressed autophagy may have a beneficial effect on the development of vitrified-warmed oocytes.
Using Atg7d/d oocytes as a negative control, we established that LC3 immunofluorescence staining can be reliably used to monitor autophagy in oocytes. While small LC3 puncta were observed in Atg7d/d oocytes at the background level, the number of large LC3 puncta was significantly higher in Atg7f/f oocytes (Figure 2), making it possible to distinguish autophagic activation by this method.
In conclusion, we propose that mouse oocytes express the Atg7 gene, and this gene is required for normal fertilization and development to the blastocyst stage but not necessary for a vitrification-warming-induced response that is autophagic.
Acknowledgments
The authors would like to thank the members of the laboratory for their constant support.
Notes
This study was supported by a grant from the Korea Health Technology R&D Project, Ministry of Health & Welfare, Korea (HI12C0055).
Conflict of interest: No potential conflict of interest relevant to this article was reported.
