Transition nuclear protein 1 as a novel biomarker in patients with fertilization failure
Article information
Abstract
Objective
Although intracytoplasmic sperm injection (ICSI) is a way to deal with in vitro fertilization failure, 3% of couples still experience repeated fertilization failure after attempted ICSI, despite having sperm within normal parameters. These patients are a challenging group whose sperm cannot fertilize the egg during ICSI. Unfortunately, no test can predict the risk of fertilization failure. Phospholipase C zeta (PLCζ) and transition nuclear proteins (TNPs) are essential factors for chromatin packaging during sperm maturation. This study aimed to assess PLCζ1 and TNP1 expression in the sperm of patients with fertilization failure and the correlations among the DNA fragmentation index, PLCζ1 and TNP1 gene and protein expression, and the risk of fertilization failure.
Methods
In this study, 12 infertile couples with low fertilization rates (<25%) and complete failure of fertilization in their prior ICSI cycles despite normal sperm parameters were chosen as the case group. Fifteen individuals who underwent ICSI for the first time served as the control group. After sperm analysis and DNA fragmentation assays, quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and Western blot analyses were performed to compare the gene and protein expression of PLCζ and TNP1 in both groups.
Results
DNA fragmentation was significantly higher in the fertilization failure group. The qRT-PCR and Western blot results demonstrated significantly lower PLCζ and TNP1 gene and protein expression in these patients than in controls.
Conclusion
The present study showed that fertilization failure in normozoospermic men was probably due to deficient DNA packaging and expression of TNP1.
Introduction
Intracytoplasmic sperm injection (ICSI) improves fertilization in couples with in vitro fertilization (IVF) failure. Nonetheless, in 1% to 3% of cases, fertilization failure still occurs after ICSI cycles are attempted, despite having sperm within normal parameters [1,2]. These patients are a challenging group whose sperm cannot fertilize the egg in the ICSI process [3]. Unfortunately, no test can predict the risk of fertilization failure or eliminate its possibility [4], and the molecular pathogenesis of fertilization failure remains unknown.
Fertilization failure after ICSI has been chiefly attributed to defects in oocyte activation [5,6]. One of the primary sperm-dependent factors for oocyte activation is phospholipase C zeta1 (PLCζ1) [7]. Other oocyte activation factors stimulate this protein to promote successful fertilization [8]. PLCζ1 is mainly expressed in the equatorial region of normal sperm and is a key factor in the complex signaling pathway that triggers Ca2+ oscillations and oocyte activation during normal fertilization [9,10]. It has been reported that PLCζ1 mutations or variations may lead to male infertility and fertilization failure due to the inability to activate oocytes [11-13]. However, the genetic basis of fertilization failure during ICSI requires further investigation. The maturity of sperm chromatin plays an important role in DNA integrity and in successful fertilization, as well as in embryo quality and development [14-16]. Multiple factors such as oxidative stress, temperature change, and environmental pollution can affect the sperm chromatin structure [17,18]. Chromatin integrity in sperm occurs during the second phase of spermiogenesis [7]. Transition nuclear protein 1 (TNP1) is the intermediate protein in the process of replacing histones with protamine and is a male-specific factor essential in sperm nuclear shaping and chromatin packaging during spermatogenesis [19,20]. Low levels of TNP1 gene expression impair normal spermatogenesis [21]. Since molecular evaluation can provide more information about sperm quality and its ability to fertilize the egg, we looked for new factors or molecules that could be used to identify these patients before they underwent the ICSI procedure [4,22]. In this study, we assessed PLCζ and TNP1 expression levels in the sperm of patients with fertilization failure and defined the associations among DNA fragmentation index (DFI), PLCζ1 and TNP1 gene and protein expression, and successful fertilization and embryo development.
Methods
1. Patient selection
This experimental study was performed at the Shahid Akbar Abadi Hospital IVF center, Iran University of Medical Sciences, Tehran, Iran. Twelve infertile couples with low-fertilization rates (<25%) and total fertilization failure in their previous ICSI cycles and normal sperm parameters according to the World Health Organization (WHO) guideline [23] were selected as a case group. Fifteen subjects who were undergoing their first ICSI trial served as a control group.
The control group included patients whose infertility was due to a tubal factor or an unexplained cause, and who were attempting ICSI treatment for the first time. They were subsequently excluded if they had a low fertilization rate (<80%) in this cycle.
In both groups, the female partners were aged 18 to 35 years. Patients who had any condition associated with severe gynecological diseases, such as polycystic ovary syndrome, endometriosis, uterine cavity abnormality, or recurrent miscarriage were excluded from the study. The exclusion criteria for the male participants in both groups included abnormal semen analysis, varicocele, and any suspected endocrine disorder.
2. Semen analysis
The semen samples were collected by masturbation after 2 to 3 days of sexual abstinence into sterile containers that were nontoxic for spermatozoa. Based on the WHO guidelines, semen analyses including sperm count, motility, and morphology were performed by an embryologist and a lab technician after sample liquefaction at room temperature for 20 minutes. Total sperm concentration and sperm motility including progressive motility, nonprogressive motility, and the percentage of immotile sperm were evaluated using a Makler chamber (under ×400 magnification). Sperm morphology was assessed using a Diff-Quick staining kit (Avicenna Laboratories Inc.). The prepared slides were evaluated by light microscopy (×100).
3. Analysis of sperm DNA fragmentation
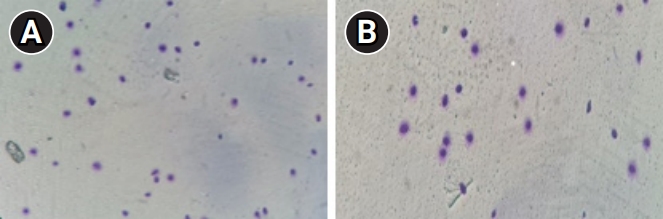
A sperm chromatin dispersion test was performed using a DNA fragmentation assay kit (Dianbio Assay Co.) to evaluate sperm DNA fragmentation (SDF). Briefly, a portion of each sample was diluted in phosphate-buffered saline to a concentration of 5 to 10×106/mL. Completely melted agarose (at 37 °C) was mixed with the sperm and placed on the supercoated slide. The slides were dipped into a denaturing solution and then into a lysing solution. After washing with distilled water, the samples were dehydrated in an ethanol series (70%, 90%, 100%). At least 200 spermatozoa per sample were evaluated at ×100 magnification. Scoring was based on halo sizes; the absence of halos or tiny halos in the spermatozoa indicated high DNA fragmentation. Finally, the percentage of SDF was reported for each sample, and a sperm DFI ≤30% was considered normal [24,25].
4. RNA extraction and cDNA synthesis
A portion of each sample was used to determine the gene expression levels of PLCζ and TNP1 by quantitative real-time polymerase chain reaction (qRT-PCR) analysis. Total RNA from the semen samples (sperm) of patients with recurrent ICSI failure and the control group were extracted using RNX-Plus (GeneAll) according to the manufacturer's protocol. The integrity of the extracted RNA and its concentration were evaluated by agarose electrophoresis and measurement of the absorbance ratio at 260 to 280 nm. Complementary DNA (cDNA) synthesis was performed using 1 μg of total RNA with the RevertAid First Strand cDNA Synthesis Kit (Bonbiotech).
5. Quantitative real-time PCR analysis
The PCR mixture for each reaction contained 7.5 μL SYBR premix Ex Taq II (Ampliqon), 1 μL of each primer (3 pmol/μL), and 12.5 ng cDNA adjusted to a final volume of 20 μL using dH2O. All reactions were performed in triplicate. Specific primer pairs were designed by Gene Runner 6.0 (Informer Technologies Inc.) (Table 1). The quantitative reverse-transcription PCR protocol included 15 minutes at 95 °C followed by 40 repeated cycles of 10 seconds at 95 °C and 35 seconds at 60 °C. The expression levels of PLCζ1 and TNP1 mRNA were normalized to the expression of the housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), for each sample. The relative expression calculations were evaluated using the ΔΔCT method [26,27].
6. Western blots
From each sample, 50 µg of protein was separated on sodium dodecyl sulfate polyacrylamide gel and then blotted onto polyvinylidene fluoride membranes. The membranes were blocked with 5% bovine serum albumin and 0.1% Tween-20. Subsequently, the membranes were incubated with the primary anti-TNP1 antibody (1:100; orb325877; Biorbyt) and GAPDH (1:200; GTX100118) overnight at 4 °C. The blots were then washed with Tris-buffered saline and polysorbate 20 (TBST) solution and incubated with horseradish peroxidase-conjugated secondary antibody (1:200; secondary [rabbit]: BA1054) (Boster Biological Technology) for 2 hours at room temperature. Immunodetection was performed after washing with TBST. The enhanced chemiluminescent peroxidase substrate (ECL; Parstous) and blots were exposed on X-ray films (Fujifilm, REF 47410). Subsequently, the films were scanned using a densitometer (GS-800; Bio-Rad Laboratories Inc.) [28].
7. Statistical analysis
In the statistical analysis, means±standard deviations were calculated for continuous variables. The independent t-test and repeated-measures analysis of variance were used to compare the study variables between groups. GraphPad Prism software version 8.0 (Dotmatics) was used for data analysis. All statistical tests were performed with a two-tailed design, and statistical significance was indicated by a p-value <0.05.
8. Compliance with ethical standards
This study was approved by the Ethics Committee of Iran University of Medical Sciences (IR.IUMS.FMD.REC.1398.179), and all participants provided written informed consent prior to sample collection.
Results
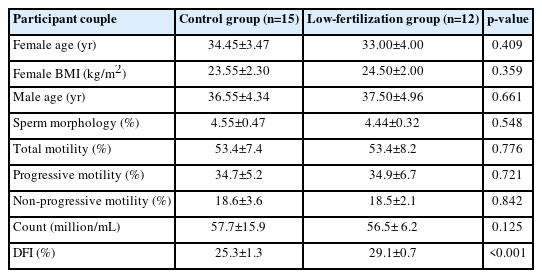
The demographic data of the participant couples are summarized in Table 2. No significant differences were found between the two groups for age and body mass index. The ejaculation volume was not significantly different between the control and fertilization failure groups. Semen macroscopic values (liquefaction, appearance, viscosity, agglutination, and aggregation) were analyzed, and no significant differences were found between the two groups (p>0.05).
1. DNA fragmentation
There was no significant difference in sperm parameters (count, motility, and morphology) between the fertilization failure group and the control group (p>0.05). However, the DFI was significantly higher in the fertilization failure group than in the controls (Table 2, Figure 1).
2. Expression of PLCζ1 and TNP1
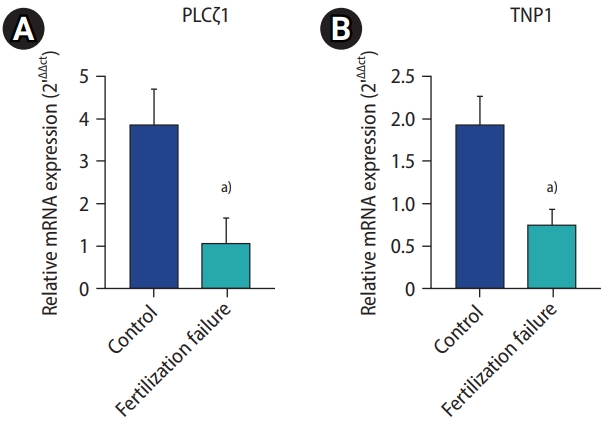
PLCζ1 and TNP1 mRNA expression levels were significantly lower in the sperm of men with recurrent fertilization failure than in the control group, according to the real-time PCR results (Figure 2).
3. Immunoblotting analysis of TNP1 protein
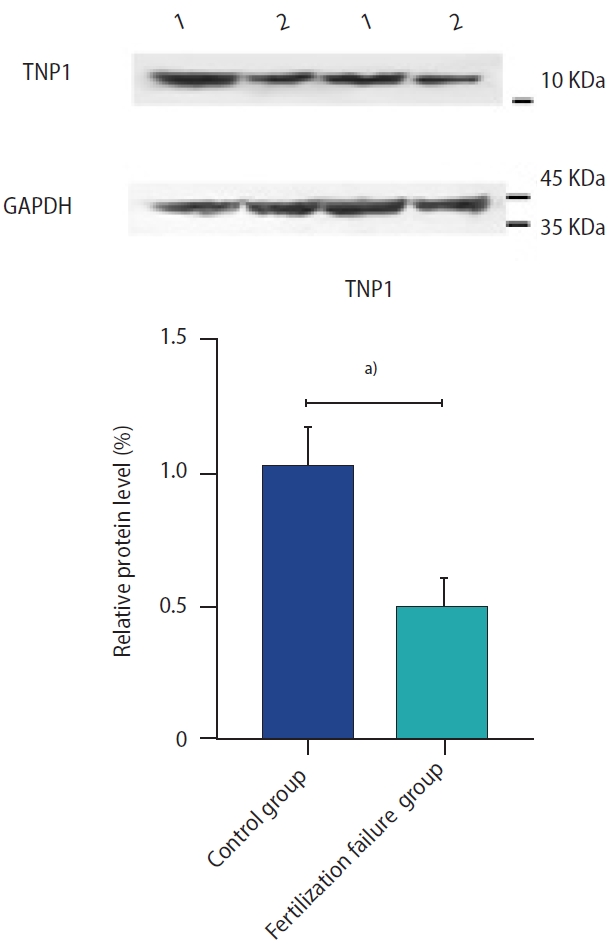
The TNP1 protein expression in sperm specimens is shown in Figure 3. Western blot analysis revealed a significant reduction in TNP1 protein expression in the sperm of men with recurrent fertilization failure as compared to the control group.
Discussion
In the present study, we assessed the mRNA and protein expression of PLCζ1 (as an index of the potential to induce oocyte fertilization) and TNP1 (as an effective factor in DNA integrity) in fertilization-failure cases and controls. Our results indicated that PLCζ1 and TNP1 were expressed at significantly lower levels in the sperm of men with fertilization failure than in the control group. Although ICSI is an accepted way to address IVF failure, some cases still experience repeated fertilization failure after attempting ICSI, despite having normal sperm parameters. No marker can predict the risk of fertilization failure, and the molecular mechanism underlying the pathogenesis remains unknown [4]. Routine semen evaluation based on the WHO protocol does not provide sufficient information to precisely predict male fertility.
Increasing evidence suggests that the integrity of sperm DNA might be the most critical factor in sperm reproductive potential, as it directly predicts oocyte fertilization. This issue has greater importance in patients with a history of fertilization failure, who typically show higher sperm DFI [29]. The compressed packaging of sperm DNA has been shown to preserve sperm genome integrity [30,31], and sperm DNA integrity plays a critical role in capacitation and acrosome reaction. It is also a key factor in the transmission of sperm genetic materials during oocyte fertilization [32,33]. When sperm chromatin damage is above 10%, oocyte repair activity is reduced [34]. Studies have shown that sperm with good morphology and high DFI values were associated with lower fertilization and pregnancy rates [15,35]. The present study showed a negative correlation between DFIs >15% and fertilization rates in the fertilization failure group. The replacement of histones by transition proteins and then by protamine plays a significant role in sperm nuclear condensation and the integrity of protection nuclear DNA [36]. Defects in protamine and TNP1 have been shown to cause DNA injury in azoospermia and varicocele patients, exacerbating infertility problems [37].
We found that the level of TNP1 gene and protein expression was significantly lower in men with recurrent fertilization failure, while abnormal sperm chromatin integrity increased the risk of fertilization failure. Thus, despite normal semen analysis, the disruption of DNA structure and TNP1 gene expression negatively affected fertilization in these patients. Therefore, it has been suggested that assessing sperm DFI and TNP1 expression may predict the fertilization potential of sperm samples in assisted reproductive technology (ART) cycles [20,35]. In line with previous reports, our results also show that the expression of PLCζ1 was significantly lower in the fertilization-failure group. Gene mutation or decreases in PLCζ1 protein abundance can result in fertilization failure, even in ICSI cycles. Egg activation by PLCζ1 initiates the embryo development process [38]. The release of PLCζ1 by sperm induces Ca2+ oscillations via the inositol 1,4,5-trisphosphate (InsP3) signaling pathway [39]. Several clinical reports have emphasized PLCζ1 deficiency in certain forms of male infertility [40,41]. Based on our findings, lower PLCζ1 and TNP1 levels may be linked with abnormal DNA integrity and a decreased rate of fertilization. The evaluation of DFI and TNP1 levels in sperm could help embryologists identify patients with null or poor fertilization outcomes before starting ART cycles. In addition, applying optimal sperm preparation techniques for microinjection to obtain high-quality sperm in terms of DNA integrity could result in higher fertilization rates and top-quality embryos. The present study had certain limitations. The authors suggest that future studies should analyze TNP1 expression in larger populations to obtain more conclusive results.
In conclusion, the present study provides novel insights into the underlying mechanisms of fertilization failure. Our results showed an association between sperm DNA damage and lower TNP1 and PLCζ expression in sperm of normozoospermic men. Abnormally low TNP1 levels can be considered a biomarker for patients with fertilization failure during ART cycles. Further investigation should be performed to clarify the clinical applications of this protein.
Notes
Conflict of interest
No potential conflict of interest relevant to this article was reported.
Author contributions
Conceptualization: FA. Data curation: ZZ, MM. Formal analysis: JSM, HG. Methodology: MGJ, MJM. Writing-original draft: JSM, HG. Writing-review & editing: FA.