Association of single-nucleotide polymorphisms in the ESR2 and FSHR genes with poor ovarian response in infertile Jordanian women
Article information
Abstract
Objective
Poor ovarian response (POR) refers to a subnormal follicular response that leads to a decrease in the quality and quantity of the eggs retrieved after ovarian stimulation during assisted reproductive treatment (ART). The present study investigated the associations of multiple variants of the estrogen receptor 2 (ESR2) and follicle-stimulating hormone receptor (FSHR) genes with POR in infertile Jordanian women undergoing ART.
Methods
Four polymorphisms, namely ESR2 rs1256049, ESR2 rs4986938, FSHR rs6165, and FSHR rs6166, were investigated in 60 infertile Jordanian women undergoing ART (the case group) and 60 age-matched fertile women (the control group), with a mean age of 33.60±6.34 years. Single-nucleotide polymorphisms (SNPs) were detected by restriction fragment length polymorphism and then validated using Sanger sequencing.
Results
The p-value of the difference between the case and control groups regarding FSHR rs6166 was very close to 0.05 (p=0.054). However, no significant differences were observed between the two groups in terms of the other three SNPs, namely ESR2 rs1256049, ESR2 rs4986938, and FSHR rs6165 (p=0.561, p=0.433, and p=0.696, respectively).
Conclusion
The association between FSHR rs6166 and POR was not statistically meaningful in the present study, but the near-significant result of this experiment suggests that statistical significance might be found in a future study with a larger number of patients.
Introduction
About 72.4 million couples suffer from infertility worldwide; accordingly, almost three million children have been conceived through assisted reproductive treatment (ART) [1]. ART is a multistep process that involves oocyte collection, oocyte fertilization, and embryo implantation [2]. The first step of ART is the collection of oocyte-containing follicles after ovarian stimulation with follicle-stimulating hormone (FSH) to obtain high-quality oocytes [3]. The response to this hormonal stimulation varies among women. Women producing 6–15 oocytes are considered normal responders, while those with not more than 4–5 oocytes are referred to as poor responders and women producing more than 15 oocytes are classified as hyperresponders [4].
Several factors, such as age, hormonal status, and ovarian reserve, play a role in the prediction of ovarian response [5,6]. In addition to previously identified predictors, various genetic polymorphisms have been proposed as markers predicting ovarian response. These variations have been observed in many genes, such as estrogen receptor 2 (ESR2) and follicle-stimulating hormone receptor (FSHR) [7,8]. It is believed that polymorphisms in the FSHR and ESR2 genes cause differences in the ovarian response and folliculogenesis [9]. FSHR is a G protein-coupled receptor (GPCR) that leads to the activation of adenylate cyclase through its main signal transduction pathway by increasing intracellular levels of cyclic adenosine monophosphate [10,11].
It is well-known that single-nucleotide polymorphisms (SNPs) in genes that play a fundamental role in oogenesis and folliculogenesis have an impact on female reproduction. To date, two different mechanisms have been proposed for this effect. Specifically, this impact can be induced by changes in the biochemical properties of a protein or at the level of transcription, which subsequently affects the activity of the promoter of a specific gene [12,13]. ESR2 and FSHR are known to influence the number of mature oocytes; therefore, they can affect the outcomes of in vitro fertilization (IVF). Boudjenah et al. [3] described that the variant of FSHR (FSHR 2039 A>G) with a G allele at position 2039 may have no effect on young people; however, it might affect people at an older age. It was also observed that patients with an A allele variant in ESR2 (ESR2 1730 G>A) had a significantly higher number of mature oocytes.
Poor ovarian response (POR) can be precisely defined as occurring when two of the three clinical criteria proposed by the European Society of Human Reproduction and Embryology (ESHRE) are present. These criteria include advanced maternal age (≥40 years), a low antral follicle count (AFC; ≤3 oocytes with conventional stimulation), and abnormal ovarian reserve test results (i.e., an anti-Müllerian hormone [AMH] level of 0.5–1.1 ng/mL) [13]. As a part of ART, gonadotropin therapy is used to stimulate ovarian function. This therapy has been reported to be successful in several aspects. With this background in mind, the present study was conducted to investigate the association of multiple variants of ESR2 and FSHR genes with POR among Jordanian women.
Methods
The current study was carried out according to the Declaration of Helsinki guidelines and approved by the Institutional Review Board of King Abdulla University Hospital in Jordan (IRB No. 2912015). Furthermore, written informed consent was obtained from all subjects or their guardians before enrollment.
1. Study population
The cohort analyzed in this study has been described in detail elsewhere [14]. To summarize, 60 female partners of selected couples undergoing ART for infertility were enrolled in the present study. The mean age of the participants was 33.60±6.34 years (range, 20–46 years). The study population was selected from couples referred to different medical centers in Jordan (King Hussein Medical Center, Islamic Hospital, Prince Rashid Hospital, and Al-Amal Maternity Hospital) to undergo controlled ovarian stimulation for IVF/intracytoplasmic sperm injection during 2014–2017. Patients with a history of endometrioma, ovarian surgery, and chemotherapy were excluded from the study.
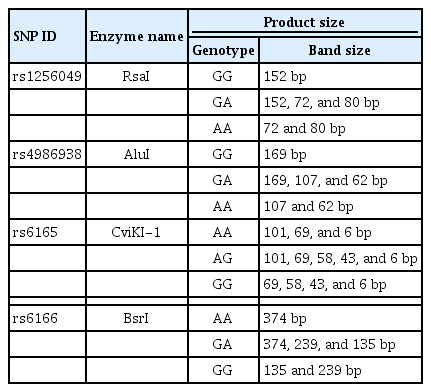
Ultrasonography was performed on the second day of the menstrual cycle to evaluate the anatomical characteristics of the female reproductive system and to identify the AFC. On the third day of the menstrual cycle, 5 mL of venous blood was collected from each participant in two tubes, including 2.5 mL in a plain tube and 2.5 mL in a tube containing tripotassium ethylenediaminetetraacetic acid (K3-EDTA). The samples in the plain tube were immediately centrifuged to separate the serum and then used for the assessment of FSH and AMH following the manufacturer’s recommendations (Beckman Coulter, San Jose, CA, USA). In addition, the blood samples in the K3-EDTA tubes were utilized to investigate the SNPs located in ESR2 and FSHR genes as shown in Table 1.
Women were included in the case group if they met two or more of the POR criteria defined by the ESHRE before the initiation of the study. The inclusion criteria were: (1) FSH level of >10 mIU/mL on the third day of the menstrual cycle, (2) AFC of <9, (3) AMH level of <1.1 ng/mL, and (4) <5 retrieved oocytes in metaphase II (MII). The subjects were divided into 10 groups based on these categories as summarized in Table 2.
In addition, 60 age-matched healthy volunteers were included in the study as controls. The participants of the control group were selected from female partners with proven fertility (i.e., with normal laboratory test results showing the potential for normal pregnancy without medical assistance). A comparison between the laboratory results of the cases and controls is presented in Table 3.
2. DNA extraction and polymerase chain reaction-restriction fragment length polymorphism detection of four SNPs
Genomic DNA was isolated from the peripheral blood samples of the case and control groups using the Gentra Puregene Blood Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The concentration and purity of the isolated DNA were measured by a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Polymerase chain reaction (PCR) was used to identify the 4 different SNPs in ESR2 and FSHR.
Two of the SNPs were located on ESR2, and the other two were on FSHR, as shown in Table 4. Each SNP was covered by its own set of primers, and all four sets of primers were designed to detect the target SNP using Primer3 Input (version 0.4.0) based on the sequences obtained from NCBI for ESR2 (NC_000014.9) and FSHR (NC_000002.12). The primers were synthesized at the Princess Haya Biotechnology Center of Jordan. Table 4 presents the primer sequences, product size, DNA variation, sequence variation, and PCR conditions for each SNP.
PCR was performed in a monoplex fashion for each primer set as indicated in Table 4. Specifically, PCR was carried out in a 0.2-mL PCR tube with a 20-μL reaction volume, containing 2 μL of template genomic DNA (~200 ng), 10 μL of 2X PCR Master Mix (New England Biolabs, Hitchin, UK), 2 μL (10 μmol) of each primer, and 20 μL of nuclease-free water. The amplification reaction for each set was conducted in a programmable thermal cycler (Thermo Fisher Scientific) as shown in Table 4. Nuclease-free water was used instead of genomic DNA as a blank to check for any DNA contamination.
The generated PCR product was run on a 2% (w/v) agarose gel prepared in 1X Tris-borate-EDTA (Sigma-Aldrich, St. Louis, MO, USA), containing ethidium bromide (Promega Corp., Madison, WI, USA). Moreover, a 50-bp DNA ladder (GeneDireX Inc., Taoyuan, Taiwan) was concurrently applied with each electrophoretic run to confirm the product size. After electrophoresis at 120 V for 45 minutes, the results were visualized and recorded using the UVP GelDoc-It2 310 Imaging System (Thermo Fisher Scientific).
Restriction fragment length polymorphism (RFLP) was carried out on the cleaned PCR products using restriction endonuclease enzymes (New England Biolabs). To determine the genotype for each sample, the PCR product was incubated with different restriction endonuclease enzymes overnight at 37°C (except for BsrI, which was incubated overnight at 65°C), as shown in Table 5. The samples were then run on a 1% agarose gel at 90 V for 1 hour. Three samples of each polymorphism with different genotypes were sent to Macrogen Inc. in South Korea to be purified and sequenced for confirmation of the RFLP results.
3. Data analysis
Patients with GG, GA, and AA alleles were considered to be normal homozygous, heterozygous, and abnormal homozygous, respectively, except for the rs6165 substitution mutation, for which subjects with AA, AG, and GG genotypes were regarded as normal homozygous, heterozygous, and abnormal homozygous, respectively. GraphPad Prism software ver. 7.0 (GraphPad, La Jolla, CA, USA) was used for the statistical analysis of Hardy-Weinberg equilibrium (HWE) [15]. All other statistical analyses were performed using version 8 of this software. The chi-square test, the Fisher exact tests (used when cells had counts of less than 5), and odds ratios (ORs) (variant homozygotes were compared to the sum of the homozygotes for the wild-type alleles plus heterozygotes) were utilized to compare differences between genotype frequencies. A p-value less than 0.05 was considered to indicate statistical significance, and 95% confidence intervals (CIs) were used to describe the strength of associations.
Results
1. Study design and clinical/demographic characteristics
A total of 60 female partners of selected couples who underwent ovarian stimulation during IVF were included in the current study. The mean levels of AMH and FSH were 0.344±0.257 ng/mL and 19.55±13.7 mIU/mL, respectively. The mean AFC and number of MII oocytes were 3.52±1.64 and 2.25+1.27 (range, 0-4) after stimulation, respectively.
2. Allele frequency distribution of the investigated genes
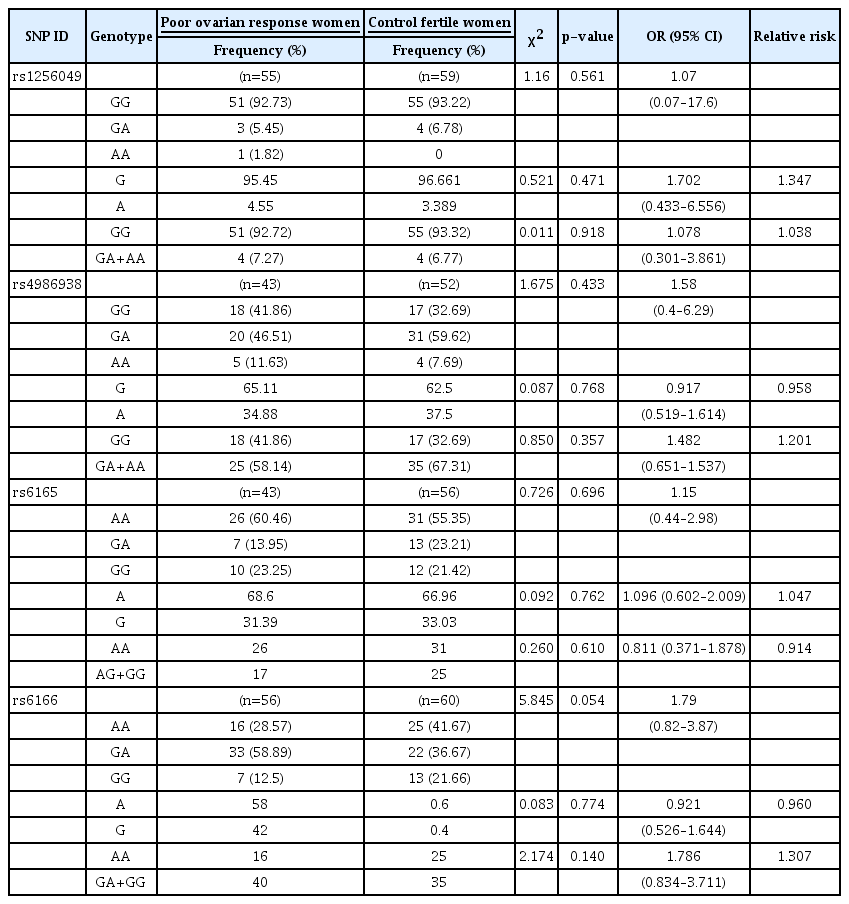
The SNPs of the two genes under investigation (ESR2 [rs1256049 and rs4986938] and FSHR [rs6165 and rs6166]) were assessed in the case and control groups to determine whether the genotype frequencies of these polymorphisms were in HWE and to investigate the association of these polymorphisms with the development of POR. Table 6 tabulates the frequency of SNP genotypes in both the case and control groups.
The investigation of the genotype frequencies of the ESR2 SNPs (rs1256049 and rs4986938) revealed that the majority of the infertile women (92.73%) were homozygotes for the wild-type allele (GG) of rs1256049, while 5.45% of the subjects were heterozygotes for the rs1256049 allele (GA) and 1.82% of the cases were homozygotes for the AA variant allele of rs1256049 (Table 6). The AA homozygous genotype was not observed in the control group. Regarding rs4986938, fewer than half of the subjects in the case (41.86%) and control (32.69%) groups were homozygotes for the wild-type allele (GG). Furthermore, 46.51% and 59.62% of the infertile and fertile women were heterozygotes for rs4986938 (GA), respectively. Additionally, 11.63% and 7.69% of the case and control groups were homozygotes for the AA variant allele of rs4986938, respectively.
Assuming random mating of the population in Jordan and applying the HWE for the distribution of the alleles, the observed genotype frequency of rs1256049 was significantly different from that predicted by HWE (p=0.001) (Table 6). However, there was no statistically significant difference among the infertile women in terms of rs4986938 (p=0.972) (Table 6). The HWE analysis of the FSHR SNPs (rs6165 and rs6166) revealed that more than half of the subjects in the case (60.46%) and control (55.35%) groups were homozygotes for the wild-type allele (AA) of rs6165. Moreover, 13.95% and 23.21% of the infertile and fertile women were heterozygotes for the rs6165 allele (AG), respectively, and 23.25% and 21.42% of the case and control groups were homozygotes for the GG variant allele of rs6165, respectively.
With regard to rs6166, fewer than a third of the infertile (28.57%) and fertile (41.67%) women were homozygotes for the wild-type allele (AA). In addition, 58.89% and 36.67% of the case and control groups were heterozygotes for the rs6166 allele (AG), respectively, and 12.50% and 21.66% of the infertile and fertile women were homozygotes for the GG variant allele of rs6166, respectively. The observed genotype frequency of rs6165 was significantly different from that expected based on HWE (p≤0.001) (Table 6) for both infertile and fertile women.
To determine significance of the associations of SNP allele and genotype frequencies with POR, the chi-square test was performed for genotypes, and p-values were calculated for each SNP. The results revealed no significant associations (p≥0.05) (Table 7), although it should be noted that rs6166 had a p-value very close to 0.05 (p=0.054) (Table 7). Furthermore, a comparison of allele frequency between the case and control groups demonstrated no significant difference in the four studied SNPs.
Association of poor ovarian response with ESR2 rs1256049 and rs4986938 and FSHR rs6165 and rs6166 alleles, and genotype frequencies
In addition, ORs were calculated for each polymorphism, with an OR of >1 indicating an association between the homozygous variant of the allele and disease. Moreover, 95% CIs were calculated to indicate how reliable the ORs were in 95% of the occasions, with a wider interval indicating greater uncertainty. The ORs calculated for rs1256049, rs4986938, rs6165, and rs6166 were 1.07 (95% CI, 0.07–17.6), 1.58 (95% CI, 0.4–6.29), 1.15 (95% CI, 0.44–2.98), and 1.79 (95% CI, 0.82–3.87), respectively (Table 7).
Discussion
It is generally believed that the outcomes of ART depend on how a woman responds to the administered gonadotropin dose. In this study, genetic variants in FSHR and ESR2 genes were investigated in infertile Jordanian women with POR and control fertile women using RFLP and Sanger sequencing (as a confirmative method). Out of the four investigated SNPs (ESR2 rs1256049, ESR2 rs4986938, FSHR rs6165, and FSHR rs6166), the p-value for the difference between the two groups regarding the rs6166 SNP in the FSHR gene was very close to 0.05 (p=0.054).
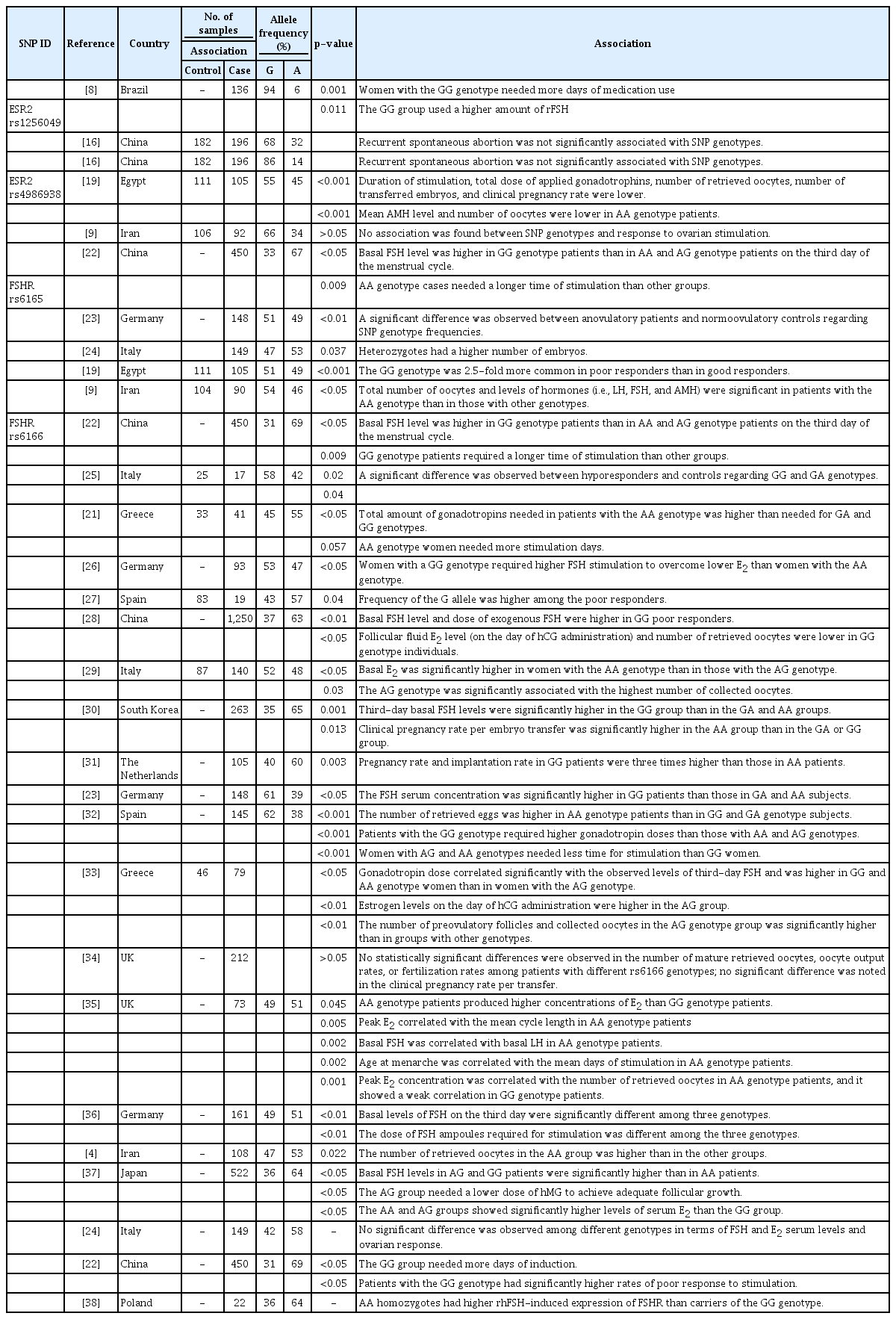
Previous studies have investigated the associations of genetic variants in ESR2 and FSHR with response to ovarian stimulation in various populations and presented variable and conflicting results. A review of the literature was carried out to identify studies that focused on the relationship of these genetic variants with various clinical parameters. Table 8 presents a summary of the collected data in this regard. Estrogen receptors (ERs) are nuclear receptors that bind to estrogen and act as transcription factors to induce follicle growth, oocyte maturation, and oocyte release, in addition to their role in uterine endometrial thickening and preparation for implantation [16,17]. Two ERs are known in humans: ERα, encoded by the ESR1 gene, and ERβ, encoded by the ESR2 gene [16,17]. Several genetic variants in ER genes have been linked to different ovarian dysfunctions [18,19]. The genetic variants of rs1256049 in ESR2 have not been extensively studied; however, this variant was reported to be associated with the amount of recombinant FSH administered and time of medication use [7]. In another study, no significant difference was observed between recurrent spontaneous abortion and rs1256049 genetic variants [20]. The rs4986938 variant occurs in a noncoding region of the ESR2 gene, and its clinical significance is not reported in ClinVar [17,18].
Summary of allele frequencies and types of associations between the four studied SNPs and clinical measurements obtained in previous studies
Two recent studies addressing the role of rs4986938 variants in ovarian response in Middle Eastern populations came to different conclusions. In a study conducted in Egypt, it was observed that women homozygous for the rs4986938 A allele variant had a lower number of retrieved oocytes after stimulation and a lower rate of clinical pregnancy [21]. However, in another study carried out in Iran, no association was reported between these SNP genotypes and response to ovarian stimulation [8]. The results of the present study are in line with the findings of the Iranian study, in which no association was observed between genetic variants in the rs4986938 SNP and ovarian response.
FSHR, a member of the GPCR family, is expressed in the granulosa cells of the ovary and is considered essential for proper FSH action [28]. The rs6165 A>G SNP is a missense variant that causes a p.Thr307Ala amino acid substitution in the FSHR protein. All published studies have confirmed the association between this genetic marker and ovarian response. Specifically, it has been demonstrated that different rs6165 genotypes were associated with basal FSH levels, duration of stimulation [30], number of obtained embryos [23], and total number of retrieved oocytes [9,19].
The rs6166 A>G SNP is another well-studied SNP in the FSHR gene that causes a p.Asn680Ser missense variation. In a number of studies, rs6166 showed an association with basal FSH levels, the time required for stimulation [21-23,28,30], the number of retrieved oocytes [4,28,29,32,39], and implantation and pregnancy rates [40]. On the contrary, in other studies, rs6166 was reported to have no association with ovarian response, especially oocyte retrieval, pregnancy rate, and FSH levels [24,34,41]. The results of the present study are consistent with the findings of the majority of previous studies regarding the important role of this SNP in determining the response of women to ovarian stimulation.
The current study was the first attempt to investigate polymorphisms in FSHR and ESR2 genes in a subset of the Jordanian Arab population. The allele and genotype frequencies were determined among women with POR and their normal counterparts. Minor allele frequencies (MAFs) were calculated for the studied SNPs in the control group, including rs1256049 MAF (A=0.039), rs4986938 MAF (A=0.375), rs6165 MAF (G=0.33), and rs6166 MAF (G=0.4). A comparison of the rs6165 MAFs obtained in this study with those reported in other studies revealed that our values were similar to those reported for populations of European and Asian origins. However, they were different from the values obtained for populations of African origin (Table 8) [25].
The rs6166 MAF was also similar to those reported for many European and Asian populations (Table 8) [42,43]. Nevertheless, only 1 study could be found regarding the Middle Eastern Arab population, which was conducted in Bahrain and reported a MAF of almost 0.5 [27]. Discrepancies among the results of various studies could be due to differences in cohort sample size, ethnicity, population stratification, and frequency of consanguinity. There are no isolated communities in Jordan; however, this country has a high rate of consanguineous marriage (20%–59%) [33]. This high consanguinity rate could explain why the genotype frequencies of the studied SNPs were out of HWE (Table 5).
The limited number of the study population and heterogeneous patients with POR are the major limitations of this study. Every year, millions of couples seek medical assistance due to infertility problems. In many ART cycles, the lack of a normal response to stimulation affects fertilization and pregnancy outcomes. Although ART is a very common therapeutic procedure in Jordan, studies on infertility and the causes of POR remain limited. The present study assessed the role of four genetic variants in two important genes, namely ESR2 and FSHR. Based on the results, only one of these variants (FSHR rs6166) should be further studied and evaluated as a marker of POR in Jordanian women. In the present study, the association between FSHR rs6166 and POR was not statistically meaningful, but the present results suggest that statistical significance may be observed in a further study with a larger number of patients.
Acknowledgements
The authors would like to extend their gratitude to Dr. Mohammed Altaleb, who is affiliated with the Department of Statistics at Yarmouk University in Jordan, for his assistance with the statistical analysis.
Notes
Conflict of interest
No potential conflict of interest relevant to this article was reported.
Author contributions
Conceptualization: AMS, OB, EHA. Data curation: OB. Formal analysis: MAH. Funding acquisition: OB. Methodology: EA, SK, GA, AA, NA, SS. Project administration: AMS, OB. Visualization: AMS, OB, EHA. Writing–original draft: AMS, OB, MAH. Writing–review & editing: AMS, OB, EHA.