 |
 |
- Search
| Clin Exp Reprod Med > Volume 50(3); 2023 > Article |
|
Abstract
Objective
Reconstructed oocytes after polar body genome transfer constitute a potential therapeutic option for patients with a history of embryo fragmentation and advanced maternal age. However, the rescue of genetic material from the first polar body (PB1) through introduction into the donor cytoplasm is not yet ready for clinical application.
Methods
Eighty-five oocytes were obtained following in vitro maturation (IVM) and divided into two groups: PB1 nuclear transfer (PB1NT; n=54) and control (n=31). Following enucleation and PB1 genomic transfer, PB1 fusion was assessed. Subsequently, all fused oocytes underwent intracytoplasmic sperm injection (ICSI) and were cultured in an incubator under a time-lapse monitoring system to evaluate fertilization, embryonic morphokinetic parameters, and cleavage patterns.
Results
Following enucleation and fusion, 77.14% of oocytes survived, and 92.59% of polar bodies (PBs) fused. However, the normal fertilization rate was lower in the PB1NT group than in the control group (56.41% vs. 92%, p=0.002). No significant differences were observed in embryo kinetics between the groups, but a significant difference was detected in embryo developmental arrest after the four-cell stage, along with abnormal cleavage division in the PB1NT group. This was followed by significant between-group differences in the implantation potential rate and euploidy status. Most embryos in the PB1NT group had at least one abnormal cleavage division (93.3%, p=0.001).
Both chromosomal abnormalities and organelle or mitochondrial dysfunction contribute to poor-quality oocytes [1]. These low-quality oocytes can result in embryo fragmentation and delayed embryo development [2], particularly in patients with advanced maternal age [3]. Nuclear transfer, a therapeutic technique, has the potential to improve oocyte quality by introducing new mitochondrial sources into the oocytes, thereby enhancing mitochondrial function and adenosine triphosphate production while minimizing the risk of aneuploidy [4]. The reconstructed oocytes contain nuclear DNA material from the patient and cytoplasm predominantly from the donor oocyte [5]. Nuclear transfer can also be utilized to prevent the transmission of mitochondrial DNA (mtDNA) mutations, treat infertility in cases of advanced maternal age or poor responders, and generate embryonic stem cells [6]. The germinal vesicle , meiotic spindle (MS), or first polar body (PB1) from matured oocytes, as well as the pronuclei (PN) of zygotes, can be used to transfer the female genomes of oocytes [7].
PB1 primarily consists of bivalent chromosomes, with a minimal presence of cytoplasm and mitochondria [7]. As a result, cytoplasm transmission to the host cytoplasm can be effectively minimized [8]. Moreover, mtDNA heteroplasmy has been detected in generations derived from oocytes reconstructed using the PB1 nuclear transfer (PB1NT) method [9]. In 1998, Wakayama and Yanagimachi [10] reported normal offspring from mouse oocytes after reconstruction and PB1NT; however, limited information is available regarding the safety and efficacy of these techniques in human embryos [11,12].
Noninvasive time-lapse monitoring (TLM) is employed at high temporal resolutions to analyze the morphokinetic events underlying developmental processes within 5 to 6 days of embryonic development [13,14]. This technique aids in calculating the time intervals between cell divisions and distinguishing between chromosomally normal and abnormal embryos by detecting multipolar divisions and abnormal nuclear structures, such as multinucleation [13,15]. Evaluating embryo development after genome transfer without blastomere removal can identify chromosomal abnormalities while maintaining the functional integrity of the embryo. Consequently, TLM can be utilized to assess the long-term safety and efficacy of human embryo genome transfer. In this study, PB1NT was used to reconstruct human embryos from oocytes derived from in vitro maturation (IVM), after which their developmental potential and cleavage patterns were evaluated. To our knowledge, morphokinetic patterns of embryos derived from reconstructed oocytes have not previously been addressed in the literature.
This study was conducted at the Yazd Institute of Reproductive Sciences. The protocol received approval from the ethics committee of Yazd University of Medical Sciences (IR.NIMAD.REC.1397.211). All patients participating in this study were informed about the research methods, and written consent was obtained. The inclusion criteria for this study were patients under 35 years old in couples affected by male factor infertility. A total of 262 immature oocytes were collected from 201 stimulated cycles.
This study was designed based on the requirement of having at least two mature oocytes with normal morphology available on the same day for genome transfer in the PB1NT group. Two oocytes with normal morphology were then allocated to the control group.
Following IVM, morphological evaluation and MS visualization of the oocytes were conducted. Oocytes displaying normal morphology, characterized by a spherical structure surrounded by a uniform zona pellucida, clear cytoplasm without inclusions, and an intact PB1 of appropriate size, were selected. These oocytes were then divided into PB1NT and control groups. The sample size was determined with the aim of having equal numbers of two-pronuclear (2PN) oocytes in each group.
IVM-derived mature oocytes, divided into two groups, were injected with sperm obtained from donor samples exhibiting normal parameters. In the PB1NT group, intracytoplasmic sperm injection (ICSI) was performed after genome transfer. Subsequently, the morphokinetic data of the injected oocytes were assessed using TLM. The specific exclusion criteria for oocytes are outlined in Figure 1.
Cumulus-corona denudation was conducted through enzymatic digestion of the cumulus complex using hyaluronidase (80 IU/mL; SAGE, CooperSurgical) and mechanical pipetting. Immature oocytes were obtained after antagonist protocol stimulation and cultured in 25-µL drops of SAGE 1-step medium (Origio; CooperSurgical) supplemented with 0.75 IU follicle-stimulating hormone and 0.75 IU luteinizing hormone (Ferring GmbH), then covered with mineral oil (LifeGlobal). The oocytes were incubated at 37 °C with 6% CO2 and 5% O2. Oocyte maturation was assessed using a stereomicroscope (Olympus) 24 hours after IVM. Mature oocytes were identified by the extrusion of the first PB into the perivitelline space [16].
The mature oocytes were placed in a 5-μL droplet of SynVitro Flush medium (Origio, CooperSurgical), which was covered by mineral oil in a glass-bottomed culture dish (WillCo-Dish; Bellco Glass). Since MS is sensitive to temperature fluctuations, extreme care was taken to maintain the oocyte at 37 °C during manipulation. MS visualization was conducted at ×200 magnification using LC Polscope optics, in conjunction with a computerized image analysis system (OCTAX PolarAIDE; OCTAX Microscience GmbH).
IVM oocytes were placed in 25-μL droplets of SynVitro Flush medium supplemented with 5 μg/mL of cytochalasin B (cat #C6762; Sigma-Aldrich) in a glass-bottomed culture dish for 5 to 10 minutes before MS removal. The MS is typically located beneath the oocyte membrane and near PB1 Figure 2A. A laser pulse was used to create a hole 13 to 15 μm in diameter in the zona pellucida adjacent to the PB1 and MS (OCTAX Laser Shot; MTG), while attempting to preserve a thin portion of the inner zona pellucida layer. Subsequently, the PB1 was removed using an enucleation pipette. For each oocyte, the PB1 was extracted with the enucleation pipette near the 3 o’clock position (Figure 2B).
The pipette, with an internal diameter of 15 to 18 μm, was filled with 15% polyvinyl pyrrolidone (PVP) for improved manipulation and suction. The MS was visualized using a PolScope imaging system and removed using the enucleation pipette without piercing the plasma membrane (Figure 2C). A normal PB was considered to have a round or oval shape with a smooth membrane surface, while fragmented PBs were discarded. PB1NT was performed when at least two mature oocytes were obtained in a single day, and PBs were transferred between the two oocytes in a crosswise manner. In this way, each oocyte served as both a donor and a recipient of the PB in the experimental group. The PB1 from another oocyte in the PB1NT group was removed Figure 2D and placed into inactivated Sendai virus extract (since inactive strand RNA virus can enhance the fusion of lipid vesicles and the cell membrane) for approximately 10 seconds (Figure 2E). Subsequently, it was placed in the perivitelline space of the enucleated oocyte (Figure 2F). The enucleated oocyte was fused with the PB1 from another oocyte in the PB1NT group. The reconstructed oocytes were inseminated via ICSI after 1 hour [17]. Finally, 2PN formation and cleavage divisions of the injected oocyte were monitored in TLM.
The oocytes were individually transferred into 5-µL droplets of SynVitro Flush medium in a glass-bottomed culture dish covered with mineral oil. Fresh ejaculated spermatozoa were prepared and placed into a central droplet of 10% PVP solution. The prepared fresh partner sperm samples were placed in a central droplet of 10% PVP solution (SAGE). The oocytes were injected under an inverted microscope with ×400 magnification (TE300; Nikon) and a heated stage at 37 °C. The injected oocytes were washed twice and cultured in droplets of SAGE 1-step medium for subsequent analysis [18].
The wells of the EmbryoSlide (Vitrolife) were filled with 25 μL of SAGE 1-step medium and covered with 1.5 mL of mineral oil. Injected oocytes were individually loaded in each well, and the dish was positioned in the EmbryoScope (Vitrolife). This advanced technology captured images every 10 minutes at seven focal planes [19]. The recorded morphokinetic parameters included the time of the second polar body extrusion (tSPBE), time of pronuclear appearance, time of pronuclear fading, and times of the first division to 2–(t2), 3–(t3), 4–(t4), 5–(t5), 6–(t6), 7–(t7), and 8–(t8). Furthermore, the duration of events in the second cell cycle (CC2a=t2−tSPBE; CC2b=t3−t2) and the second synchrony (S2=t4−t3) were also reported. Additionally, abnormal cleavage patterns, such as direct cleavage (the cleavage of a single cell into three or four blastomeres) and reverse cleavage (the reduction in the number of blastomeres due to cell fusion), were observed [14].
Additionally, uneven blastomeres were noted, in which the average diameter of the larger blastomere was more than 20% greater than that of the smaller blastomere. Significant fragmentation was observed, characterized by two daughter blastomeres accompanied by a large fragment or 10% to 50% scattered fragments following division. Micronuclei were defined as smaller versions of primary nuclei containing only one or a few chromosomes [13]. Distorted cytoplasmic movement was documented, which involved a series of irregular cytoplasmic movements occurring during or after cell division and before the embryo entered a quiescent state. Disordered division was also recorded, in which the division of one blastomere from the previous cleavage was delayed until after the other blastomere had completed the subsequent division.
A predictive evaluation of implantation potential and euploidy status in embryos was conducted using algorithms developed by Basile et al. [20] for those that reached the five-cell stage. Embryo categorization in terms of implantation potential was based on three time points—t3, CC2, and t—from the Basile algorithm. In this algorithm, embryos were categorized from A to D, with the highest implantation potential rate observed in grade A embryos (32%) [20]. Embryo categorization in terms of euploidy status was determined based on two aneuploidy markers: t5−t2 and CC3. In this algorithm, embryos were also categorized from A to D, with grade A embryos having the greatest chance of being chromosomally normal [21].
Statistical analysis was performed using SPSS version 20 (IBM Corp.). The Kolmogorov-Smirnov test was used to assess normal distribution. Results were reported as mean±standard deviation for all variables. t-tests and Mann-Whitney U tests were employed to determine the p-value differences between groups for variables with normal and non-normal distributions, respectively. Categorical variables were compared using the chi-square and Fisher exact tests. p-values of <0.05 were considered to indicate statistical significance.
A total of 262 immature oocytes were collected after denudation (Figure 1). Following maturation and evaluation, 125 metaphase II (MII) oocytes were included in the study. Of these, 77.14% (n=54) of the IVM-derived oocytes survived after enucleation and were used for genome transfer. A fusion rate of 92.59% (50 oocytes) was detected, determined by the absence of PB in the perivitelline space. After ICSI, 39 (72.2%) reconstructed oocytes were fertilized, but only 22 of them formed 2PN zygotes (56.41%). The remaining oocytes exhibited abnormal fertilization patterns (mono-pronuclear [1PN] or three-pronuclear [3PN]). In the control group, 25 of 31 oocytes were fertilized (80.6%), and 23 of the 25 fertilized oocytes were normal, displaying 2PN (92%). No significant difference was observed between the two groups in total fertilization rate (p=0.38). However, significant differences were detected in 2PN and 1PN formation rates between the two groups (p=0.003 and p=0.023, respectively). Details of the data after ICSI are provided in Table 1. Total fertilization refers to the sum of 1PN, 2PN, and 3PN. Normal fertilization and abnormal fertilization are reflected by 2PN and 1PN+3PN, respectively.
To assess developmental competency, all normal zygotes were cultured for 5 days under TLM. Although three blastocysts formed in the control group, none of the embryos in the PB1NT group reached the blastocyst stage. Consequently, the comparative evaluation of embryonic developmental potential was based on the time points obtained by the end of the third cleavage division. As shown in Table 2, the kinetic data analysis revealed no significant differences in any resulting time point between the two groups (p>0.05) (Table 2). Additionally, fewer embryos successfully completed their cleavage divisions on the third day in the PB1NT group (n=1) than in the control group (n=6), as shown in Figure 3A. Regardless of time point, the images indicated that 93.33% of embryos exhibited at least one abnormal behavior during the cleavage stage in the PB1NT group. This rate was significantly higher than that in the control group. Notable differences were observed between the two groups in terms of micronucleation, distorted cytoplasmic movement, and reverse cleavage rates (Figure 3B).
Based on Basile’s revised embryo categorization algorithm, direct cleavage, uneven blastomere, and multinucleation at day 2 were considered exclusion criteria, and embryos with these features were omitted. Ultimately, eight and nine embryos (in the PB1NT and control groups, respectively) were compared in terms of implantation potential and euploidy status based on the Basile algorithm (Table 3). In the control group, 44.44% of the embryos were categorized as “A” regarding implantation potential rate, compared to 12.5% in the PB1NT group. Furthermore, more than half of the embryos were categorized as “D” in the PB1NT group. According to the Basile algorithm, 90% of these embryos may have abnormal chromosomal statuses. In contrast, in the control group, a higher proportion of embryos were classified as “A” with respect to euploid conditions (Table 3).
Genome transfer techniques, such as germinal vesicle, spindle transfer, PB, and PN approaches, are considered suitable options for reconstructing oocyte cytoplasm containing few or dysfunctional mitochondria. These methods are particularly useful in cases involving poor-quality oocytes, inherited mitochondrial diseases, and age-related cytoplasmic deficiencies in advanced maternal age [9,11,22]. As a counterpart to the spindle chromosomal complex, PB1 contains the same copy number variation and homologous chromosomal content as the oocyte nucleus, with intact microtubules [9]. However, a limited number of studies have examined the efficacy of this technique in human oocytes.
Regarding oocyte survival rates after enucleation and normal fertilization rates through micromanipulation and PB1NT, our findings are consistent with previous studies involving reconstruction of human oocytes using PB1NT [11,12]. Similarly, Ma et al. [12] reported lower blastocyst formation rates in PB1NT, which aligns with our observations that PB1NT embryos did not reach the blastocyst stage in the present study. While Ma et al. [12] obtained MII oocytes from healthy volunteers aged 25 to 31 years, we utilized rescue IVM of immature oocytes. Previous research has confirmed that the number of transcripts in the gene expression profile is significantly reduced in in vivo MII oocytes [23], which may contribute to the decreased developmental potential of these oocytes [24,25].
The present research revealed no significant differences between the two groups regarding cleavage time intervals; however, the PB1NT group exhibited more abnormal kinetic patterns of cleavage and poorer embryonic development. In the PB1NT group, 59.09% of embryos were arrested before the four-cell stage. An evaluation of the implantation potential of embryos using the Basile algorithm revealed that only 12.5% of the obtained embryos in the PB1NT group reached the five-cell stage and were categorized as A+. In contrast, 66.66% of embryos in the control group were scored as top quality (grade A+ or B). The remaining embryos (87.5%) in the PB1NT group were categorized as D [20].
At least one abnormal cleavage pattern and nuclear structure was observed in 14 out of 15 embryos in the PB1NT group. Basile et al. [21] introduced two discrete time points to distinguish chromosomally normal embryos from abnormal ones. This hierarchical classification of embryos was based on t5−t2 >20.5 hours per CC3: 11–18 hours [21]. In the present study, these two time points were found to be outside the optimal range in 62.5% of the embryos in the PB1NT group, which were categorized as grade D. Basile et al. [21] reported abnormal chromosomal conditions in approximately 90% of these embryos. The differences in implantation potential and euploidy may account for the variations in embryonic development observed between the two groups.
A direct relationship exists between chromosomal defects and embryo morphology, specifically in terms of scattered fragmentation and multinucleation [24]. Micronucleation [26] and chromosomal sequestration through cellular fragments [13] are involved in aneuploidy and poor embryonic development, which is associated with natural negative selection against aneuploidy [27,28]. The single embryo classified as A+ in terms of potential implantation in the PB1NT group was categorized as D in terms of euploidy.
Research indicates that the coinheritance of maternal nuclear and mtDNA is essential for functional interactions between nuclear and mitochondrial gene products [29,30]. The introduction of low levels of heteroplasmy to human oocytes during nuclear transfer leads to genomic drift of mitochondria [31], which compromises mitochondrial function and results in abnormal chromosomal development and embryo arrest. Notably, despite oocyte manipulation, no synchronization was observed between cytoplasmic and nuclear development in the PB1NT group compared to the control group [6].
Challenges of the PB1NT technique include the potential for oocyte degeneration during the extrusion of the MS, as the genome is aspirated while enveloped by a small fraction of cytoplasm. Additionally, fusion is performed using either the hemagglutinating virus of Japan envelope vector or an electrical pulse, which can cause premature exit from meiosis [32]. The yet-unknown effect of the vector on the oocyte can be partially explained through clinical trials [33]. However, incomplete discharge of the MS may occur, potentially increasing the likelihood of 3PN formation. In the present study, we relied on the appearance of PB to select those with an intact genome. Similarly, Ma et al. [12] reported a higher 1PN formation in the PB1NT group. Given that none of the 1PN-derived reconstructed oocytes failed PB2 extrusion, this indicates that the non-responsive donor genome in the host cytoplasm contributed to 1PN formation [12], and it is likely that the transplanted PB is not viable. Unfortunately, no method exists for assessing PB viability, and the use of certain stains, such as Hoechst dye, should not be applied in clinical treatment. Furthermore, the safety of using cytochalasin requires further investigation.
Although the reconstructed oocytes in this study underwent successful fertilization and embryo formation, we observed lower developmental potential in cleavage embryos derived from these reconstructed oocytes compared to the control group, accompanied by abnormal cleavage patterns.
Acknowledgments
This study was part of a PhD thesis and was financially supported by the National Institute for Medical Research Development (NIMAD, No. 971155), Tehran, Iran.
We would like to thank the staff of the laboratory of reproductive medicine.
Figure 1.
Recruitment flow chart of 169 in vitro maturation oocyte. PB1NT, first polar body (PB1) nuclear transfer.
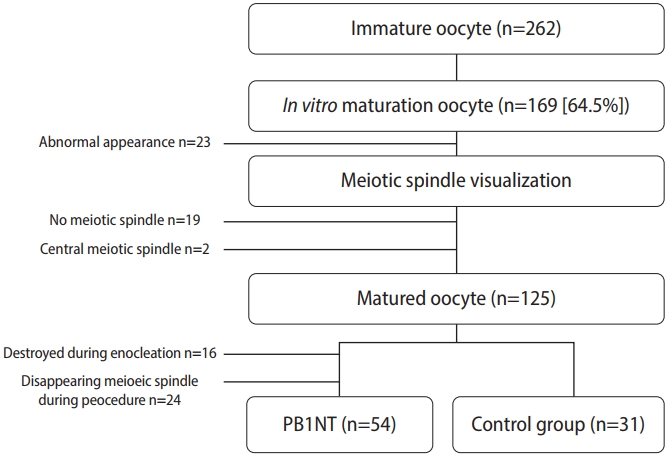
Figure 2.
Experimental design for serial polar body (PB) transfer between two oocytes, ×200 magnification. (A) Matured oocyte with a visible meiotic spindle (by PolScope, OCTAX PolarAID; OCTAX) from first PB nuclear transfer (PB1NT) group (arrow 1: meiotic spindle; arrow 2: PB). (B) Piercing the zona with a laser (OCTAX Laser Shot; MTG) and removing the PB1 oocyte. (C) Isolation of the meiotic spindle (arrow). (B) Biopsy of PB1 from another oocyte in PB1NT and using it as a genome donor. (E) Donor PB1 placed in hemagglutinating virus of Japan Envelope Vector. (F) Donor PB1 placed in the sub-zona environment.

Figure 3.
(A) Comparison of embryonic developmental potential between control and first polar body nuclear transfer (PB1NT) groups. (B) The proportion and distribution of abnormal behavior during early embryonic development by time-lapse imaging. PN, pronuclei; DC, direct cleavage; RC, reverse cleavage; DD, distorted cleavage; BF, big fragmentation; DCM, distorted cytoplasmic movement. a)p<0.0001. The p-values were calculated using the chi-square test or Fisher exact test.
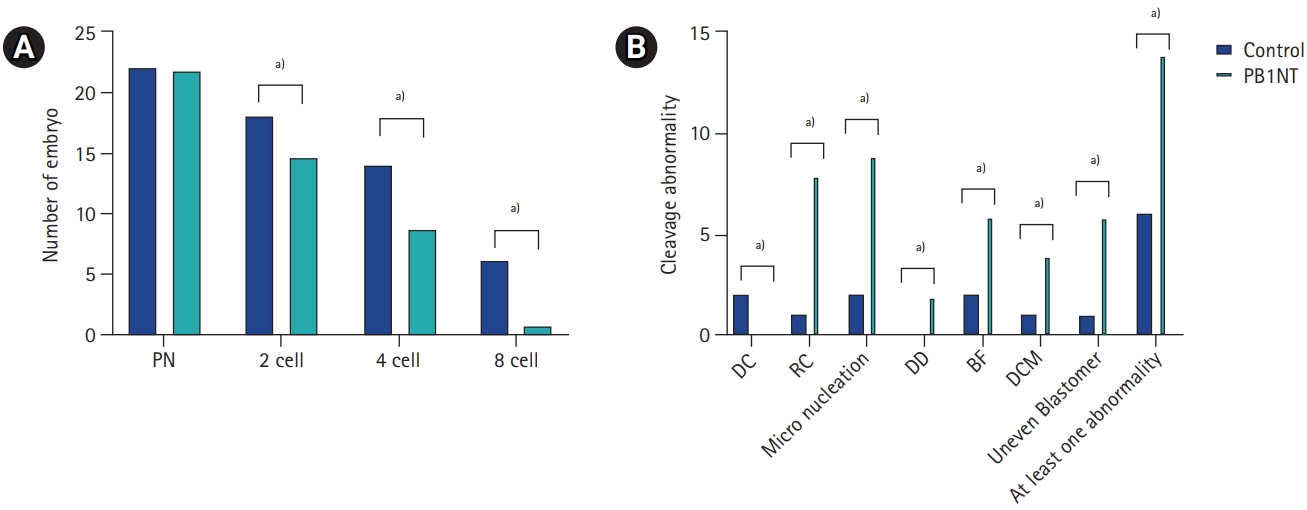
Table 1.
Differences in fertilization rates between the PB1NT and control groups
Table 2.
Comparison of the kinetic data of cleavage embryos between two groups
Values are presented as mean±standard deviation.
PB1NT, first polar body (PB1) nuclear transfer; tSPBE, time of the second PB extrusion; tPNA, time of pronuclei appearance; tPNF, time of pronuclei fading; t2, time of 2-cell stage; CC1, t2–tPNf; t3, time of 3-cell stage; t4, time of 4-cell stage; CC2a, t3–t2; CC2b, t4–t2; t5, time of 5-cell stage; t6, time of 6-cell stage; t7, time of 7-cell stage; t8, time of 8-cell stage.
a)p-value calculated with the t-test; b)Calculated with Mann-Whitney U test.
Table 3.
Comparative evaluation of embryo implantation potential and euploidy status between the two groups
References
1. Igarashi H, Takahashi T, Nagase S. Oocyte aging underlies female reproductiveaging: biological mechanisms and therapeutic strategies. Reprod Med Biol 2015;14:159-69.




2. Balaban B, Urman B. Effect of oocyte morphology on embryo development and implantation. Reprod Biomed Online 2006;12:608-15.


3. Cimadomo D, Fabozzi G, Vaiarelli A, Ubaldi N, Ubaldi FM, Rienzi L. Impact of maternal age on oocyte and embryo competence. Front Endocrinol (Lausanne) 2018;9:327.



4. Labarta E, de Los Santos MJ, Escriba MJ, Pellicer A, Herraiz S. Mitochondria as a tool for oocyte rejuvenation. Fertil Steril 2019;111:219-26.


5. Amato P, Tachibana M, Sparman M, Mitalipov S. Three-parent in vitro fertilization: gene replacement for the prevention of inherited mitochondrial diseases. Fertil Steril 2014;101:31-5.



6. Zhang J, Zhuang G, Zeng Y, Grifo J, Acosta C, Shu Y, et al. Pregnancy derived from human zygote pronuclear transfer in a patient who had arrested embryos after IVF. Reprod Biomed Online 2016;33:529-33.


7. Ou XH, Sun QY. Mitochondrial replacement techniques or therapies (MRTs) to improve embryo development and to prevent mitochondrial disease transmission. J Genet Genomics 2017;44:371-4.


8. Wu K, Zhong C, Chen T, Zhang X, Tao W, Zhang J, et al. Polar bodies are efficient donors for reconstruction of human embryos for potential mitochondrial replacement therapy. Cell Res 2017;27:1069-72.




9. Wang T, Sha H, Ji D, Zhang HL, Chen D, Cao Y, et al. Polar body genome transfer for preventing the transmission of inherited mitochondrial diseases. Cell 2014;157:1591-604.


10. Wakayama T, Yanagimachi R. The first polar body can be used for the production of normal offspring in mice. Biol Reprod 1998;59:100-4.


11. Zhang SP, Lu CF, Gong F, Xie PY, Hu L, Zhang SJ, et al. Polar body transfer restores the developmental potential of oocytes to blastocyst stage in a case of repeated embryo fragmentation. J Assist Reprod Genet 2017;34:563-71.




12. Ma H, O'Neil RC, Marti Gutierrez N, Hariharan M, Zhang ZZ, He Y, et al. Functional human oocytes generated by transfer of polar body genomes. Cell Stem Cell 2017;20:112-9.



13. Daughtry BL, Chavez SL. Time-lapse imaging for the detection of chromosomal abnormalities in primate preimplantation embryos. Methods Mol Biol 2018;1769:293-317.


14. Faramarzi A, Khalili MA, Ashourzadeh S, Palmerini MG. Does rescue in vitro maturation of germinal vesicle stage oocytes impair embryo morphokinetics development? Zygote 2018;26:430-4.


15. Aguilar J, Rubio I, Munoz E, Pellicer A, Meseguer M. Study of nucleation status in the second cell cycle of human embryo and its impact on implantation rate. Fertil Steril 2016;106:291-9.


16. Fesahat F, Dehghani Firouzabadi R, Faramarzi A, Khalili MA. The effects of different types of media on in vitro maturation outcomes of human germinal vesicle oocytes retrieved in intracytoplasmic sperm injection cycles. Clin Exp Reprod Med 2017;44:79-84.




17. Tachibana M, Sparman M, Mitalipov S. Chromosome transfer in mature oocytes. Nat Protoc 2010;5:1138-47.




18. Omidi M, Khalili MA, Ashourzadeh S, Rahimipour M. Zona pellucida birefringence and meiotic spindle visualisation of human oocytes are not influenced by IVM technology. Reprod Fertil Dev 2014;26:407-13.


19. Roesner S, Dietrich JE, Weigert J, Montag M, Toth B, Strowitzki T. Time-lapse imaging reveals differences in growth dynamics of embryos after in vitro maturation compared with conventional stimulation. Fertil Steril 2017;107:606-12.


20. Basile N, Vime P, Florensa M, Aparicio Ruiz B, Garcia Velasco JA, Remohi J, et al. The use of morphokinetics as a predictor of implantation: a multicentric study to define and validate an algorithm for embryo selection. Hum Reprod 2015;30:276-83.


21. Basile N, Nogales Mdel C, Bronet F, Florensa M, Riqueiros M, Rodrigo L, et al. Increasing the probability of selecting chromosomally normal embryos by time-lapse morphokinetics analysis. Fertil Steril 2014;101:699-704.


22. Wolf DP, Mitalipov N, Mitalipov S. Mitochondrial replacement therapy in reproductive medicine. Trends Mol Med 2015;21:68-76.



23. Jones GM, Cram DS, Song B, Magli MC, Gianaroli L, Lacham-Kaplan O, et al. Gene expression profiling of human oocytes following in vivo or in vitro maturation. Hum Reprod 2008;23:1138-44.


24. Magli MC, Ferraretti AP, Crippa A, Lappi M, Feliciani E, Gianaroli L. First meiosis errors in immature oocytes generated by stimulated cycles. Fertil Steril 2006;86:629-35.


25. Nogueira D, Staessen C, Van de Velde H, Van Steirteghem A. Nuclear status and cytogenetics of embryos derived from in vitro-matured oocytes. Fertil Steril 2000;74:295-8.


26. Liu S, Kwon M, Mannino M, Yang N, Renda F, Khodjakov A, et al. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature 2018;561:551-5.




27. Mantikou E, Wong KM, Repping S, Mastenbroek S. Molecular origin of mitotic aneuploidies in preimplantation embryos. Biochim Biophys Acta 2012;1822:1921-30.


28. Maurer M, Ebner T, Puchner M, Mayer RB, Shebl O, Oppelt P, et al. Chromosomal aneuploidies and early embryonic developmental arrest. Int J Fertil Steril 2015;9:346-53.


29. Dunham-Snary KJ, Ballinger SW. Genetics. Mitochondrial-nuclear DNA mismatch matters. Science 2015;349:1449-50.



30. Reinhardt K, Dowling DK, Morrow EH. Medicine. Mitochondrial replacement, evolution, and the clinic. Science 2013;341:1345-6.


31. Yamada M, Egli D. Genome transfer prevents fragmentation and restores developmental potential of developmentally compromised postovulatory aged mouse oocytes. Stem Cell Reports 2017;8:576-88.



- TOOLS







