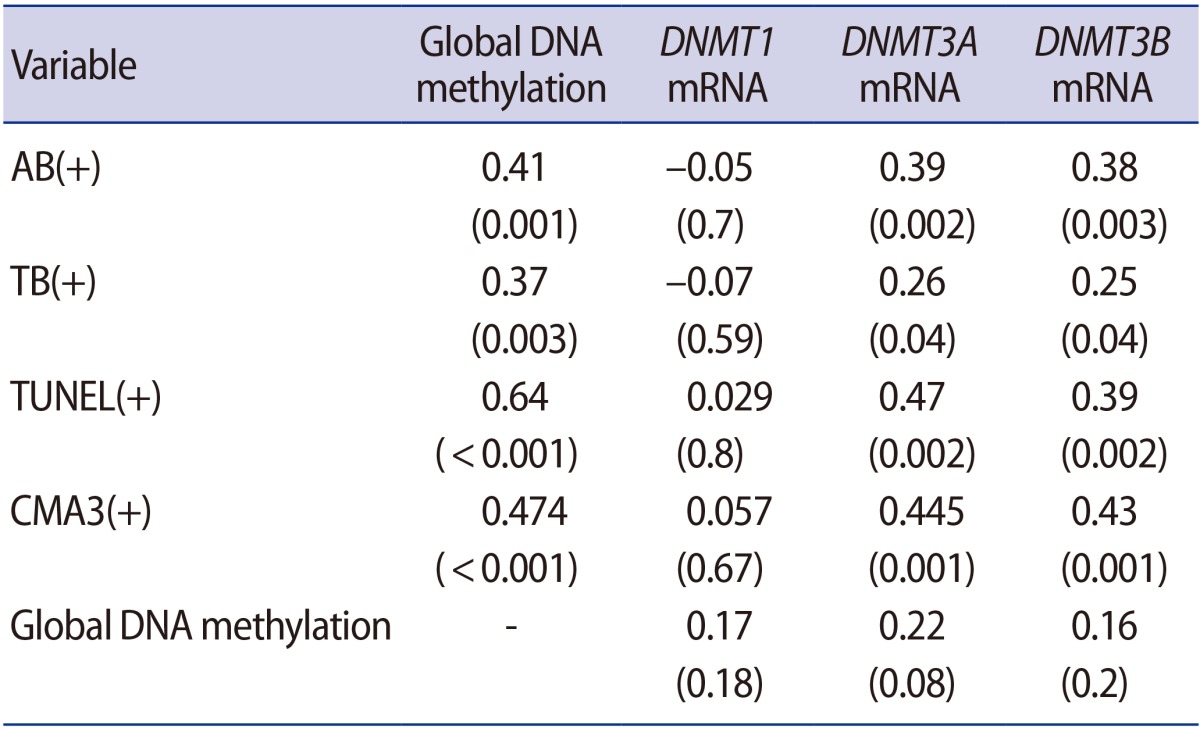
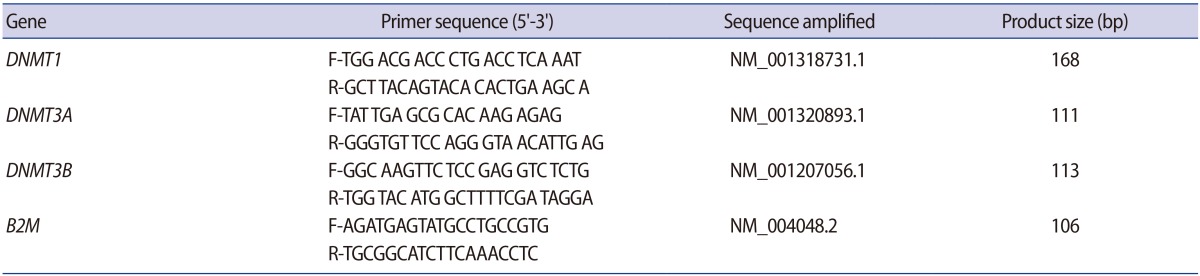
1. Cavallini G. Male idiopathic oligoasthenoteratozoospermia. Asian J Androl 2006;8:143-157.PMID:
16491265.


3. Cui X, Jing X, Wu X, Yan M, Li Q, Shen Y, et al. DNA methylation in spermatogenesis and male infertility. Exp Ther Med 2016;12:1973-1979.PMID:
27698683.



4. Camprubi C, Pladevall M, Grossmann M, Garrido N, Pons MC, Blanco J. Semen samples showing an increased rate of spermatozoa with imprinting errors have a negligible effect in the outcome of assisted reproduction techniques. Epigenetics 2012;7:1115-1124.PMID:
22885410.



5. Yanagisawa Y, Ito E, Yuasa Y, Maruyama K. The human DNA methyltransferases DNMT3A and DNMT3B have two types of promoters with different CpG contents. Biochim Biophys Acta 2002;1577:457-465.PMID:
12359337.


6. Messerschmidt DM, Knowles BB, Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev 2014;28:812-828.PMID:
24736841.



7. Kato Y, Nozaki M. Distinct DNA methylation dynamics of spermatogenic cell-specific intronless genes is associated with CpG content. PLoS One 2012;7:e43658PMID:
22952732.



8. Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, Siegmund KD, et al. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet 2012;131:1565-1589.PMID:
22740325.



10. Li L, Le F, Wang LY, Xu XR, Lou HY, Zheng YM, et al. Normal epigenetic inheritance in mice conceived by in vitro fertilization and embryo transfer. J Zhejiang Univ Sci B 2011;12:796-804.PMID:
21960342.



11. Daxinger L, Whitelaw E. Transgenerational epigenetic inheritance: more questions than answers. Genome Res 2010;20:1623-1628.PMID:
21041414.



12. Latham KE, Schultz RM. Embryonic genome activation. Front Biosci 2001;6:D748-D759.PMID:
11401780.


13. Aoki VW, Emery BR, Carrell DT. Global sperm deoxyribonucleic acid methylation is unaffected in protamine-deficient infertile males. Fertil Steril 2006;86:1541-1543.PMID:
17011559.


14. Olszewska M, Barciszewska MZ, Fraczek M, Huleyuk N, Chernykh VB, Zastavna D, et al. Global methylation status of sperm DNA in carriers of chromosome structural aberrations. Asian J Androl 2017;19:117-124.PMID:
26908061.


15. Benchaib M, Braun V, Ressnikof D, Lornage J, Durand P, Niveleau A, et al. Influence of global sperm DNA methylation on IVF results. Hum Reprod 2005;20:768-773.PMID:
15640258.



16. Menkveld R. Clinical significance of the low normal sperm morphology value as proposed in the fifth edition of the WHO Laboratory Manual for the Examination and Processing of Human Semen. Asian J Androl 2010;12:47-58.PMID:
20111081.



17. Talebi AR, Khalili MA, Hossaini A. Assessment of nuclear DNA integrity of epididymal spermatozoa following experimental chronic spinal cord injury in the rat. Int J Androl 2007;30:163-169.PMID:
17298547.


18. Rosenborg L, Rao KM, Bjorndahl L, Kvist U, Pousette A, Akerlof E, et al. Changes in human sperm chromatin stability during preparation for in-vitro fertilization. Int J Androl 1990;13:287-296.PMID:
2387649.


19. Talebi AR, Moein MR, Tabibnejad N, Ghasemzadeh J. Effect of varicocele on chromatin condensation and DNA integrity of ejaculated spermatozoa using cytochemical tests. Andrologia 2008;40:245-251.PMID:
18727735.


20. Agarwal A, Bragais FM, Sabanegh E. Assessing sperm function. Urol Clin North Am 2008;35:157-171.PMID:
18423237.


21. Rahiminia T, Hosseini A, Anvari M, Ghasemi-Esmailabad S, Talebi AR. Modern human sperm freezing: effect on DNA, chromatin and acrosome integrity. Taiwan J Obstet Gynecol 2017;56:472-476.PMID:
28805603.


22. Talebi AR, Khalili MA, Vahidi S, Ghasemzadeh J, Tabibnejad N. Sperm chromatin condensation, DNA integrity, and apoptosis in men with spinal cord injury. J Spinal Cord Med 2013;36:140-146.PMID:
23809529.



23. Talebi AR, Vahidi S, Aflatoonian A, Ghasemi N, Ghasemzadeh J, Firoozabadi RD, et al. Cytochemical evaluation of sperm chromatin and DNA integrity in couples with unexplained recurrent spontaneous abortions. Andrologia 2012;44(Suppl 1): 462-470.PMID:
21806662.


24. Nallella KP, Sharma RK, Aziz N, Agarwal A. Significance of sperm characteristics in the evaluation of male infertility. Fertil Steril 2006;85:629-634.PMID:
16500330.


25. World Health Organization. WHO laboratory manual for the examination and processing of human semen. Geneva: World Health Organization; 2010.
26. Agarwal A, Said TM. Role of sperm chromatin abnormalities and DNA damage in male infertility. Hum Reprod Update 2003;9:331-345.PMID:
12926527.



27. Shamsi MB, Imam SN, Dada R. Sperm DNA integrity assays: diagnostic and prognostic challenges and implications in management of infertility. J Assist Reprod Genet 2011;28:1073-1085.PMID:
21904910.



28. Sakkas D, Moffatt O, Manicardi GC, Mariethoz E, Tarozzi N, Bizzaro D. Nature of DNA damage in ejaculated human spermatozoa and the possible involvement of apoptosis. Biol Reprod 2002;66:1061-1067.PMID:
11906926.



29. Marques CJ, Joao Pinho M, Carvalho F, Bieche I, Barros A, Sousa M. DNA methylation imprinting marks and DNA methyltransferase expression in human spermatogenic cell stages. Epigenetics 2011;6:1354-1361.PMID:
22048249.


30. La Salle S, Trasler JM. Dynamic expression of DNMT3a and DNMT3b isoforms during male germ cell development in the mouse. Dev Biol 2006;296:71-82.PMID:
16725135.


31. Cheng P, Chen H, Zhang RP, Liu SR, Zhou-Cun A. Polymorphism in DNMT1 may modify the susceptibility to oligospermia. Reprod Biomed Online 2014;28:644-649.PMID:
24631383.


32. Pacheco SE, Houseman EA, Christensen BC, Marsit CJ, Kelsey KT, Sigman M, et al. Integrative DNA methylation and gene expression analyses identify DNA packaging and epigenetic regulatory genes associated with low motility sperm. PLoS One 2011;6:e20280PMID:
21674046.



33. Yu B, Zhou H, Liu M, Zheng T, Jiang L, Zhao M, et al. Epigenetic alterations in density selected human spermatozoa for assisted reproduction. PLoS One 2015;10:e0145585PMID:
26709917.



34. Barzideh J, Scott RJ, Aitken RJ. Analysis of the global methylation status of human spermatozoa and its association with the tendency of these cells to enter apoptosis. Andrologia 2013;45:424-429.PMID:
23121197.


35. Aoki VW, Emery BR, Liu L, Carrell DT. Protamine levels vary between individual sperm cells of infertile human males and correlate with viability and DNA integrity. J Androl 2006;27:890-898.PMID:
16870950.


36. Novakovic B, Wong NC, Sibson M, Ng HK, Morley R, Manuelpillai U, et al. DNA methylation-mediated down-regulation of DNA methyltransferase-1 (DNMT1) is coincident with, but not essential for, global hypomethylation in human placenta. J Biol Chem 2010;285:9583-9593.PMID:
20071334.



37. Kitamura A, Miyauchi N, Hamada H, Hiura H, Chiba H, Okae H, et al. Epigenetic alterations in sperm associated with male infertility. Congenit Anom (Kyoto) 2015;55:133-144.PMID:
26212350.


38. Zama AM, Uzumcu M. Epigenetic effects of endocrine-disrupting chemicals on female reproduction: an ovarian perspective. Front Neuroendocrinol 2010;31:420-439.PMID:
20609371.